Multiple myeloma pathophysiology: Difference between revisions
No edit summary |
m (Bot: Removing from Primary care) |
||
| (278 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{Multiple myeloma}} | {{Multiple myeloma}} | ||
{{CMG}} | {{CMG}} {{AE}} {{HMHJ}}; {{HL}}; {{shyam}} | ||
== | |||
==Overview== | |||
[[Multiple myeloma]], a disorder of clonal late [[B-cells]], arises from post-[[Germinal centers|germinal center]] [[plasma cells]] that are normally involved in production of human [[Immunoglobulins|immunoglobulins.]]<ref name="radio">Multiple myeloma. Radiopaedia (2015)http://radiopaedia.org/articles/multiple-myeloma-1 Accessed on September, 20th 2015</ref><ref name="wiki">Multiple myeloma. Wikipedia (2015)https://en.wikipedia.org/wiki/Multiple_myeloma#Pathophysiology Accessed on September, 20th 2015</ref><ref name="Med">Multiple myeloma. Medlineplus (2015)https://www.nlm.nih.gov/medlineplus/multiplemyeloma.html Accessed on September, 20th 2015</ref> Although the exact [[pathogenesis]] and the stage at which [[Multiple myeloma|myeloma cells]] arise from post-[[Germinal centers|germinal]] [[B-cells]] remain unclear, a variety of factors have been implicated in [[pathogenesis]] of [[multiple myeloma]]. Of these, [[Chromosome|chromosomal]] abnormalities are thought to be the most important. It has been suggested that all cases of [[multiple myeloma]] pass through [[MGUS]]. [[Kidney|Renal]] involvement by [[multiple myeloma]] is catergorized into three entities: [[Light chain nephropathy|light chain cast nephropathy]], [[monoclonal]] [[immunoglobulin]] deposition disease, and [[amyloidosis]]. [[Bone|Osseous]] involvement by [[multiple myeloma]] is based on [[cytokine]] and [[cellular]] interactions that lead to [[Bone cell|bone]] breakdown. On microscopic histopathological analysis, abundant [[eosinophilic]] [[cytoplasm]], eccentrically placed [[nucleus]], and ''[[Russell bodies]]'' are characteristic findings of [[multiple myeloma]].<ref name="patho">Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015</ref> | |||
==Pathophysiology== | ==Pathophysiology== | ||
=== | ===Normal physiology and development of plasma cells=== | ||
* [[Plasma cells]] are terminally differentiated [[B-cells]] which function to produce [[immunoglobulins]]. [[plasma cells]] arise from [[B lymphocytes]] through a series of events. These events take place in [[bone marrow]] and secondary [[Lymphoid organs|lymphoid tissues]]. | |||
* These events include [[immunoglobulin]] [[Heavy-chain immunoglobulin|heavy-chain]] (IgH) VDJ [[gene]] rearrangement, migration into [[Lymphoid tissue|lymphoid tissues]], [[somatic hypermutation]] (SMH) in the [[Immunoglobulin heavy chain|IgH]] and [[Immunoglobulin light chains|light-chain]] [[Gene|genes]], [[antigen]] selection, switch in Ig class from [[IgM]] to [[IgG]], [[IgA]], [[IgD]], or [[IgE]], migration back to [[bone marrow]] and terminal [[differentiation]] into [[Plasma cell|plasma cells]].<ref name="pmid23176722" /> <ref name="pmid18070707" /><ref name="pmid25723853" /> | |||
'''Stem cells → Pre-B cells → Immature B-cells → Mature B-cells (naïve) → Activated B-cells → Memory B-cells and Plasmablasts → Plasma cells''' | |||
* During these events, [[B cells]] express variety of surface markers which may be used to denote their [[developmental]] stage such as [[CD19]], [[CD20]], [[CD27]], [[CD38]], [[CD10]], [[CD138]]. These markers and [[Immunoglobulin heavy chain|IgH]] chain [[gene]] sequences are important to define the nature of [[Multiple myeloma|myeloma]] cells.<ref name="pmid23176722" /><ref name="pmid18070707" /><ref name="pmid25723853" /> | |||
* [[Plasma cells]] secrete [[antibodies]] which function in [[humoral immunity]].<ref name="pmid23176722" /><ref name="pmid18070707" /> <ref name="pmid25723853" /> | |||
* [[Plasma cell|Plasma cells]] are typically [[polyclonal]] and can respond to a variety of [[antigens]], which helps combat [[infections]]. Under normal circumstances, there is no [[Monoclonal|monoclonality]] amongst the [[plasma cell]] population in a person. These [[plasma cells]] are functionally intact in their ability to contribute to [[humoral immunity]].<ref name="pmid23176722">{{cite journal |vauthors=Borrello I |title=Can we change the disease biology of multiple myeloma? |journal=Leuk. Res. |volume=36 Suppl 1 |issue= |pages=S3–12 |date=November 2012 |pmid=23176722 |pmc=3698609 |doi=10.1016/S0145-2126(12)70003-6 |url=}}</ref><ref name="pmid18070707">{{cite journal |vauthors=Chng WJ, Glebov O, Bergsagel PL, Kuehl WM |title=Genetic events in the pathogenesis of multiple myeloma |journal=Best Pract Res Clin Haematol |volume=20 |issue=4 |pages=571–96 |date=December 2007 |pmid=18070707 |pmc=2198931 |doi=10.1016/j.beha.2007.08.004 |url=}}</ref><ref name="pmid25723853">{{cite journal |vauthors=Hengeveld PJ, Kersten MJ |title=B-cell activating factor in the pathophysiology of multiple myeloma: a target for therapy? |journal=Blood Cancer J |volume=5 |issue= |pages=e282 |date=February 2015 |pmid=25723853 |pmc=4349256 |doi=10.1038/bcj.2015.3 |url=}}</ref> | |||
* Transcription factors such as interferon regulatory factor 4 (IRF4), BCL6, B-lymphocyte-induced maturation protein 1 (BLIMP1, also known as PRDM1), paired box gene 5 (PAX5) and X box binding protein 1 (XBP1) play an important role in differentiation and survival of plasma cells.<ref name="pmid24688108">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid11460154">{{cite journal |vauthors=Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH |title=Plasma cell differentiation requires the transcription factor XBP-1 |journal=Nature |volume=412 |issue=6844 |pages=300–7 |date=July 2001 |pmid=11460154 |doi=10.1038/35085509 |url=}}</ref><ref name="pmid21924923">{{cite journal |vauthors=Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD |title=The genetic network controlling plasma cell differentiation |journal=Semin. Immunol. |volume=23 |issue=5 |pages=341–9 |date=October 2011 |pmid=21924923 |doi=10.1016/j.smim.2011.08.010 |url=}}</ref> | |||
'''Interferon regulatory factor 4 (IRF4) → Down-regulation of BCL6 → Up-regulation of B-lymphocyte-induced maturation protein 1 (BLIMP1) → Down-regulation of paired box gene 5 (PAX5) and Up-regulation of X box binding protein 1 (XBP1).'''<ref name="pmid246881082">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid114601542">{{cite journal |vauthors=Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH |title=Plasma cell differentiation requires the transcription factor XBP-1 |journal=Nature |volume=412 |issue=6844 |pages=300–7 |date=July 2001 |pmid=11460154 |doi=10.1038/35085509 |url=}}</ref><ref name="pmid219249232">{{cite journal |vauthors=Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD |title=The genetic network controlling plasma cell differentiation |journal=Semin. Immunol. |volume=23 |issue=5 |pages=341–9 |date=October 2011 |pmid=21924923 |doi=10.1016/j.smim.2011.08.010 |url=}}</ref> | |||
* | |||
For more information on [[plasma cells]], [[Plasma cell|click here]]. | |||
=== Normal physiology and development of Immunoglobulins === | |||
* '''Immunoglobulins''' (also known as '''Antibodies''') are [[protein]]s that are found in [[blood]] or other [[bodily fluid]]s of [[vertebrate]]s, and are used by the [[immune system]] to identify and neutralize foreign objects, such as [[bacterium|bacteria]] and [[virus]]es. | |||
* They are made of a few basic structural units called ''chains''; each antibody has two large [[heavy chains]] '''H''' and two small [[light chain]]s '''L'''. There are several different types of [[antibody]] [[heavy chains]], and several different kinds of [[antibodies]], which are grouped into different ''[[isotype (immunology)|isotypes]]'' based on which heavy chain they possess.<ref name="Market" /><ref name="pmid8450761" /> | |||
* Five different antibody isotypes are known in mammals, which perform different roles, and help direct the appropriate immune response for each different type of foreign object they encounter.<ref name="Market">Eleonora Market, F. Nina Papavasiliou (2003) [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0000016 ''V(D)J Recombination and the Evolution of the Adaptive Immune System''] [[PLoS Biology]]1(1): e16.</ref><ref name="pmid8450761">{{cite journal |author=Litman GW, Rast JP, Shamblott MJ, ''et al'' |title=Phylogenetic diversification of immunoglobulin genes and the antibody repertoire |journal=Mol. Biol. Evol. |volume=10 |issue=1 |pages=60–72 |year=1993 |pmid=8450761 |doi=}}</ref> | |||
* [[Class switch recombination]] (CSR), a region specific deletion recombination process, generates different [[immunoglobulin]] (Ig) isotypes by replacing one switch region with another. This process leads to enhanced functionality of [[immunoglobulins]].<ref name="pmid246881083">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid219249233">{{cite journal |vauthors=Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD |title=The genetic network controlling plasma cell differentiation |journal=Semin. Immunol. |volume=23 |issue=5 |pages=341–9 |date=October 2011 |pmid=21924923 |doi=10.1016/j.smim.2011.08.010 |url=}}</ref> | |||
* [[Class switch recombination]] (CSR), just like [[somatic hypermutation]] (SMH), requires the [[expression]] of activation-induced deaminase (AID) and both are dependent on creation of double-stranded [[DNA]] breaks (DSB) in the [[Immunoglobulin|Ig]] [[loci]].<ref name="pmid246881084">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid23307864">{{cite journal |vauthors=Keim C, Kazadi D, Rothschild G, Basu U |title=Regulation of AID, the B-cell genome mutator |journal=Genes Dev. |volume=27 |issue=1 |pages=1–17 |date=January 2013 |pmid=23307864 |pmc=3553278 |doi=10.1101/gad.200014.112 |url=}}</ref><ref name="pmid17634408">{{cite journal |vauthors=González D, van der Burg M, García-Sanz R, Fenton JA, Langerak AW, González M, van Dongen JJ, San Miguel JF, Morgan GJ |title=Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma |journal=Blood |volume=110 |issue=9 |pages=3112–21 |date=November 2007 |pmid=17634408 |doi=10.1182/blood-2007-02-069625 |url=}}</ref> | |||
For more information on [[immunoglobulins]], [[Immunoglobulin|click here]]. | |||
===Pathogenesis=== | |||
The [[pathogenesis]] of [[multiple myeloma]] is complex and probably is a result of multiple and multi-step [[oncogenic]] events such as hyperdiploidy and deregulation of ''[[cyclin D1]]'', and interaction of [[Multiple myeloma|myeloma]] [[Cell (biology)|cells]] with [[Bone marrow|marrow]] environment. Recently it has been suggested that all cases of [[multiple myeloma]] pass through an [[MGUS]] phase. The events surrounding the progression of [[MGUS]] into [[multiple myeloma]] are not well-defined but environmental and [[Genetics|genetic]] factors have been proposed to have an association. A brief description of events thought to play a role in [[pathogenesis]] of [[multiple myeloma]] is given here. | |||
==== Biology of myeloma cells ==== | |||
* [[Multiple myeloma|Myeloma]] [[cells]] are [[malignant]] [[plasma cells]] which exhibit the [[Morphology (biology)|morphology]] of mature [[plasma cells]] or plasmablasts. Majority of these [[cells]] seem to be mature, differentiated and [[quiescent]], appearing to be without long-term proliferative potential.<ref name="pmid231767222" /><ref name="pmid14630803">{{cite journal |vauthors=Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ |title=Characterization of clonogenic multiple myeloma cells |journal=Blood |volume=103 |issue=6 |pages=2332–6 |date=March 2004 |pmid=14630803 |pmc=3311914 |doi=10.1182/blood-2003-09-3064 |url=}}</ref> | |||
* [[Cells]] with similar [[morphology]] to mature [[B-cells]] but with [[immunoglobulin]] [[Gene|gene sequences]] and [[idiotype]] similar to [[Multiple myeloma|myeloma cells]] have also been found in [[Multiple myeloma|myeloma]] [[Patient|patients]], both in [[bone marrow]] and the [[peripheral blood]].<ref name="pmid12795416" /><ref name="pmid14630803" /> | |||
* Current [[hypothesis]] is the presence of functional [[heterogeneity]] in [[Multiple myeloma|myeloma cells]] with only a minor group of specialized [[Multiple myeloma|myeloma cells]] exhibiting '''clonogenic''' growth potential. Although studies have shown the presence of [[Multiple myeloma|myeloma]] cells sub-populations with distinct [[phenotypes]] and functionality in [[multiple myeloma]] such as presence of [[CD138]]+ and [[CD138]]− sub-populations, the [[Phenotypes|phenotype]] of these so-called clonogenic [[cells]] is yet to be determined. <ref name="pmid12795416" /><ref name="pmid14630803" /><ref name="pmid18539970">{{cite journal |vauthors=Huff CA, Matsui W |title=Multiple myeloma cancer stem cells |journal=J. Clin. Oncol. |volume=26 |issue=17 |pages=2895–900 |date=June 2008 |pmid=18539970 |pmc=2610256 |doi=10.1200/JCO.2007.15.8428 |url=}}</ref> | |||
* [[Multiple myeloma|Myeloma]] cells express surface markers associated with [[plasma cells]] such as [[CD138]], [[Natural killer cells|natural killer (NK) cells]] such as [[CD56]]/NCAM, T cells such as [[CD28]], and sometimes the pan-[[B-cell]] marker [[CD20]]. Presence of [[CD19]] and [[CD20]] on sub-population of [[Multiple myeloma|myeloma cells]] may suggest either early-lineage [[Precursors|precursor]] for [[Multiple myeloma|myeloma cells]] or possible de-[[differentiation]] of [[Multiple myeloma|myeloma]] [[cells]].<ref name="pmid23176722" /><ref name="pmid14630803" /><ref name="pmid16956823">{{cite journal |vauthors=Bataille R, Jégo G, Robillard N, Barillé-Nion S, Harousseau JL, Moreau P, Amiot M, Pellat-Deceunynck C |title=The phenotype of normal, reactive and malignant plasma cells. Identification of "many and multiple myelomas" and of new targets for myeloma therapy |journal=Haematologica |volume=91 |issue=9 |pages=1234–40 |date=September 2006 |pmid=16956823 |doi= |url=}}</ref><ref name="pmid3115338">{{cite journal |vauthors=Grogan TM, Durie BG, Lomen C, Spier C, Wirt DP, Nagle R, Wilson GS, Richter L, Vela E, Maxey V |title=Delineation of a novel pre-B cell component in plasma cell myeloma: immunochemical, immunophenotypic, genotypic, cytologic, cell culture, and kinetic features |journal=Blood |volume=70 |issue=4 |pages=932–42 |date=October 1987 |pmid=3115338 |doi= |url=}}</ref><ref name="pmid10373068">{{cite journal |vauthors=Kiel K, Cremer FW, Rottenburger C, Kallmeyer C, Ehrbrecht E, Atzberger A, Hegenbart U, Goldschmidt H, Moos M |title=Analysis of circulating tumor cells in patients with multiple myeloma during the course of high-dose therapy with peripheral blood stem cell transplantation |journal=Bone Marrow Transplant. |volume=23 |issue=10 |pages=1019–27 |date=May 1999 |pmid=10373068 |doi=10.1038/sj.bmt.1701767 |url=}}</ref> | |||
* [[Myeloma]] cells show complex [[Chromosome|chromosomal]] abnormalities and [[mutations]]. Studies have demonstrated the presence of [[translocations]] in up to 75% of the [[Multiple myeloma|myeloma]] cases.<ref name="pmid23176722" /> | |||
==== Environmental and hereditary factors ==== | |||
* The Key environmental and [[hereditary]] factors thought to confer a greater risk of developing [[multiple myeloma]] or play a part in [[pathogenesis]] are summarized in the table below.<ref name="pmid24688108">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid12199621">{{cite journal |vauthors=Morgan GJ, Davies FE, Linet M |title=Myeloma aetiology and epidemiology |journal=Biomed. Pharmacother. |volume=56 |issue=5 |pages=223–34 |date=July 2002 |pmid=12199621 |doi= |url=}}</ref><ref name="pmid20713974">{{cite journal |vauthors=Wadhera RK, Rajkumar SV |title=Prevalence of monoclonal gammopathy of undetermined significance: a systematic review |journal=Mayo Clin. Proc. |volume=85 |issue=10 |pages=933–42 |date=October 2010 |pmid=20713974 |pmc=2947966 |doi=10.4065/mcp.2010.0337 |url=}}</ref><ref name="pmid19179464">{{cite journal |vauthors=Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D, Hoover R, Rajkumar SV |title=Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study |journal=Blood |volume=113 |issue=22 |pages=5412–7 |date=May 2009 |pmid=19179464 |pmc=2689042 |doi=10.1182/blood-2008-12-194241 |url=}}</ref><ref name="pmid23568533">{{cite journal |vauthors=Wang SS, Voutsinas J, Chang ET, Clarke CA, Lu Y, Ma H, West D, Lacey JV, Bernstein L |title=Anthropometric, behavioral, and female reproductive factors and risk of multiple myeloma: a pooled analysis |journal=Cancer Causes Control |volume=24 |issue=7 |pages=1279–89 |date=July 2013 |pmid=23568533 |pmc=3684420 |doi=10.1007/s10552-013-0206-0 |url=}}</ref><ref name="pmid17617273">{{cite journal |vauthors=Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM |title=Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis |journal=Lancet |volume=370 |issue=9581 |pages=59–67 |date=July 2007 |pmid=17617273 |doi=10.1016/S0140-6736(07)61050-2 |url=}}</ref><ref name="pmid23564249">{{cite journal |vauthors=Kachuri L, Demers PA, Blair A, Spinelli JJ, Pahwa M, McLaughlin JR, Pahwa P, Dosman JA, Harris SA |title=Multiple pesticide exposures and the risk of multiple myeloma in Canadian men |journal=Int. J. Cancer |volume=133 |issue=8 |pages=1846–58 |date=October 2013 |pmid=23564249 |doi=10.1002/ijc.28191 |url=}}</ref><ref name="pmid2246554">{{cite journal |vauthors=Singh J, Dudley AW, Kulig KA |title=Increased incidence of monoclonal gammopathy of undetermined significance in blacks and its age-related differences with whites on the basis of a study of 397 men and one woman in a hospital setting |journal=J. Lab. Clin. Med. |volume=116 |issue=6 |pages=785–9 |date=December 1990 |pmid=2246554 |doi= |url=}}</ref><ref name="pmid19582882">{{cite journal |vauthors=Kristinsson SY, Björkholm M, Goldin LR, Blimark C, Mellqvist UH, Wahlin A, Turesson I, Landgren O |title=Patterns of hematologic malignancies and solid tumors among 37,838 first-degree relatives of 13,896 patients with multiple myeloma in Sweden |journal=Int. J. Cancer |volume=125 |issue=9 |pages=2147–50 |date=November 2009 |pmid=19582882 |pmc=2737604 |doi=10.1002/ijc.24514 |url=}}</ref><ref name="pmid22120009">{{cite journal |vauthors=Broderick P, Chubb D, Johnson DC, Weinhold N, Försti A, Lloyd A, Olver B, Ma Y, Dobbins SE, Walker BA, Davies FE, Gregory WA, Childs JA, Ross FM, Jackson GH, Neben K, Jauch A, Hoffmann P, Mühleisen TW, Nöthen MM, Moebus S, Tomlinson IP, Goldschmidt H, Hemminki K, Morgan GJ, Houlston RS |title=Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk |journal=Nat. Genet. |volume=44 |issue=1 |pages=58–61 |date=November 2011 |pmid=22120009 |pmc=5108406 |doi=10.1038/ng.993 |url=}}</ref><ref name="pmid23955597">{{cite journal |vauthors=Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Försti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, Hose D, Walker BA, Davies FE, Gregory WA, Jackson GH, Irving JA, Pratt G, Fegan C, Fenton JA, Neben K, Hoffmann P, Nöthen MM, Mühleisen TW, Eisele L, Ross FM, Straka C, Einsele H, Langer C, Dörner E, Allan JM, Jauch A, Morgan GJ, Hemminki K, Houlston RS, Goldschmidt H |title=Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk |journal=Nat. Genet. |volume=45 |issue=10 |pages=1221–1225 |date=October 2013 |pmid=23955597 |pmc=5053356 |doi=10.1038/ng.2733 |url=}}</ref> | |||
{| class="wikitable" style="width:40%; height:100px" border="1" | |||
|+ | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Environmental and hereditary risk factors | |||
|- | |||
| | |||
* Increasing age | |||
* Gender (increased incidences in male) | |||
* [[Familial]] history | |||
* Past history of [[MGUS]] (Monoclonal gammopathy of undetermined significance) | |||
* Obesity | |||
* Immune dysfunction such as [[Auto-immune disorders|auto-immune]] [[disease]], [[Human Immunodeficiency Virus (HIV)|HIV]] ([[Human Immunodeficiency Virus|human immunodeficiency virus]]) and [[Organ transplant|transplant]] recipients | |||
* Exposure to [[chemicals]], [[pesticides]] or [[radiation]] | |||
* Greater [[prevalence]] of [[MGUS]] and [[Multiple myeloma|multiple myeloma]] in African Americans* | |||
* [[Inherited]] variation in [[genetic]] [[loci]] at 2p, 3p, 3q, 6p, 7p, 17p and 22q | |||
|- | |||
|* Likely influenced by environmental and behavioral [[Confounding factor|confounding factors]]. | |||
|} | |||
==== Chromosomal aberrations ==== | |||
* Two major sets of [[Chromosome|chromosomal]] abnormalities seen in [[multiple myeloma]] are [[translocations]] with extensive IgH rearrangements and numerical aberrations, either [[trisomy]] or [[monosomy]].<ref name="pmid24688108">{{cite journal |vauthors=Boyle EM, Davies FE, Leleu X, Morgan GJ |title=Understanding the multiple biological aspects leading to myeloma |journal=Haematologica |volume=99 |issue=4 |pages=605–12 |date=April 2014 |pmid=24688108 |pmc=3971069 |doi=10.3324/haematol.2013.097907 |url=}}</ref><ref name="pmid23176722">{{cite journal |vauthors=Borrello I |title=Can we change the disease biology of multiple myeloma? |journal=Leuk. Res. |volume=36 Suppl 1 |issue= |pages=S3–12 |date=November 2012 |pmid=23176722 |pmc=3698609 |doi=10.1016/S0145-2126(12)70003-6 |url=}}</ref> | |||
===== '''''Translocations''''' ===== | |||
* Majority of [[translocations]] in [[multiple myeloma]] take place through [[class switch recombination]] (CSR). But few may occur through D<sub>H</sub>–J<sub>H</sub> recombination, possibly in early stages of [[B-cell]] development such as pre-[[B cell]] stage.<ref name="pmid24688108" /><ref name="pmid23435460">{{cite journal |vauthors=Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, Ross FM, Davies FE, Gonzalez D, Morgan GJ |title=Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells |journal=Blood |volume=121 |issue=17 |pages=3413–9 |date=April 2013 |pmid=23435460 |doi=10.1182/blood-2012-12-471888 |url=}}</ref> | |||
* Majority of these [[translocations]] put [[oncogenes]] such as [[Cyclin-D1|cyclin]] D1 (CCND1), [[CCND3]], [[fibroblast growth factor receptor 3]] ([[FGFR3]]), multiple myeloma SET domain (MMSET; also known as WHSC1), [[MAF (gene)|MAF]] and [[MAFB (gene)|MAFB]] under the influence of [[Enhancer (genetics)|enhancers]] present at Ig [[loci]]. .<ref name="pmid23176722" /><ref name="pmid24688108" /><ref name="pmid9207791">{{cite journal |vauthors=Chesi M, Nardini E, Brents LA, Schröck E, Ried T, Kuehl WM, Bergsagel PL |title=Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3 |journal=Nat. Genet. |volume=16 |issue=3 |pages=260–4 |date=July 1997 |pmid=9207791 |pmc=3901950 |doi=10.1038/ng0797-260 |url=}}</ref><ref name="pmid19798094">{{cite journal |vauthors=Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H |title=International Myeloma Working Group molecular classification of multiple myeloma: spotlight review |journal=Leukemia |volume=23 |issue=12 |pages=2210–21 |date=December 2009 |pmid=19798094 |pmc=2964268 |doi=10.1038/leu.2009.174 |url=}}</ref> | |||
* These [[translocations]] cause [[Overexpression|over-expression]] of these [[oncogenes]] that in turn leads to dysregulation of [[Cell cycle|cell-cycle]] such as increased cell survival, growth, progression and [[DNA repair]]. One of the key abnormality being the increased [[G1/S transition]] during the [[Cell cycle|cell-cycle]].<ref name="pmid24688108" /><ref name="pmid19059936">{{cite journal |vauthors=Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, Avramidou A, Irving JA, Gonzalez D, Davies FE, Morgan GJ |title=MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells |journal=Haematologica |volume=94 |issue=1 |pages=78–86 |date=January 2009 |pmid=19059936 |pmc=2625417 |doi=10.3324/haematol.13426 |url=}}</ref> | |||
* Many other [[translocations]] may occur in later phases of [[multiple myeloma]]. One important example is [[translocation]] of MYC, an oncogene that encodes [[transcription factors]].<ref name="pmid24688108" /><ref name="pmid10618400">{{cite journal |vauthors=Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM |title=Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=97 |issue=1 |pages=228–33 |date=January 2000 |pmid=10618400 |pmc=26645 |doi= |url=}}</ref><ref name="pmid2025578">{{cite journal |vauthors=Nobuyoshi M, Kawano M, Tanaka H, Ishikawa H, Tanabe O, Iwato K, Asaoku H, Sakai A, Kuramoto A |title=Increased expression of the c-myc gene may be related to the aggressive transformation of human myeloma cells |journal=Br. J. Haematol. |volume=77 |issue=4 |pages=523–8 |date=April 1991 |pmid=2025578 |doi= |url=}}</ref> | |||
===== '''''Hyperdiploidy''''' ===== | |||
* Gain of the odd numbered [[chromosomes]] including 3, 5, 7, 9, 11, 15, 19 and 21 is the other major abnormality observed in [[multiple myeloma]].<ref name="pmid23176722" /> <ref name="pmid24688108" /> | |||
* The mechanism of this gain phenomena is not well understood, one [[hypothesis]] being the gain of [[chromosome]] during single [[mitosis]].<ref name="pmid24688108" /> | |||
{| | |||
! colspan="3" style="background:#4479BA; color: #FFFFFF;" align="center" + |<big>Chromosomal aberrations in multiple myeloma (MM)</big> | |||
|- | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + | Chromosomal Abnormality | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + | Chromosome(s)/Protein(s) affected | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + | Consequence | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |[[Trisomies]] | |||
| align="left" style="background:#F5F5F5;" + |Odd-numbered [[chromosomes]] with the exception of [[chromosomes]] 1, 13, and 21 | |||
| align="left" style="background:#F5F5F5;" + | | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(11;14)(q13;q32) | |||
t(6;14q)(p21;32) | |||
t(12;14)(p13;q32) | |||
| align="left" style="background:#F5F5F5;" + |[[Cyclin D|Cyclin]] D1 | |||
[[Cyclin D|Cyclin]] D3 | |||
[[Cyclin D|Cyclin]] D2 | |||
| align="left" style="background:#F5F5F5;" + |Over-expression; [[cell cycle]] dysregulation | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(4;14)(p16;q32) | |||
| align="left" style="background:#F5F5F5;" + |[[FGFR3]] or MMSET | |||
| align="left" style="background:#F5F5F5;" + |Over-expression and activation; [[Multiple myeloma|multiple myeloma]] cell [[proliferation]]/[[apoptosis]] prevention MMSET probably linked to crucial transforming event | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |t(14;16)(q32;q23) | |||
t(14;20)(q32;q11) | |||
t(8;14)(q24;q32) | |||
| align="left" style="background:#F5F5F5;" + |''c-[[MAF (gene)|MAF]]'' | |||
[[MAFB (gene)|MAFB]] | |||
[[MAFA]] | |||
| align="left" style="background:#F5F5F5;" + |Over-expression; involvement in [[IL-4]] regulation | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |del 17p13 | |||
| align="left" style="background:#F5F5F5;" + |[[p53]] | |||
| align="left" style="background:#F5F5F5;" + |[[Cell cycle|Cell-cycle]] dysregulation/[[apoptosis]] | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |[[Monosomy]] 14 | |||
| align="left" style="background:#F5F5F5;" + |[[Chromosome]] 14 | |||
| align="left" style="background:#F5F5F5;" + | | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |[[Chromosome]] 13 deletion and monosomy | |||
| align="left" style="background:#F5F5F5;" + |[[Chromosome]] 13 | |||
| align="left" style="background:#F5F5F5;" + | | |||
|- | |||
! align="center" style="background:#DCDCDC;" + |Gain(1q21) | |||
| align="left" style="background:#F5F5F5;" + |[[Chromosome 1|Chromosome]] 1 | |||
| align="left" style="background:#F5F5F5;" + | | |||
|- | |||
! align="center" style="background:#DCDCDC;" + colspan="3" |'''Abbreviations used:''' [[FGFR3]]:[[fibroblast growth factor receptor 3]]; MMSET:multiple myeloma SET domain; [[MAF (gene)|MAF]]:[[MAF (gene)|musculoaponeurotic fibrosarcoma oncogene homolog]]. | |||
|} | |||
==== Mutations in myeloma ==== | |||
* Studies have demonstrated the presence of approximately 35 [[mutations]] per sample in [[multiple myeloma]]. These [[mutations]] cause loss of [[Tumor suppressor|tumor suppressors]] and [[NFKB1|NFKB]] alterations.<ref name="pmid24688108" /> | |||
===== '''''Tumor suppressors''''' ===== | |||
* These [[mutations]] result in [[Cell cycle|cell-cycle]] dysregulation with an increase in [[G1/S transition]]. Some of these [[mutations]] are mentioned in the table below.<ref name="pmid24688108" /><ref name="pmid20616218">{{cite journal |vauthors=Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, Johnson DC, Gonzalez D, Dagrada GP, Protheroe RK, Konn ZJ, Stockley DM, Gregory WM, Davies FE, Ross FM, Morgan GJ |title=A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value |journal=Blood |volume=116 |issue=15 |pages=e56–65 |date=October 2010 |pmid=20616218 |doi=10.1182/blood-2010-04-279596 |url=}}</ref><ref name="pmid16840723">{{cite journal |vauthors=Gonzalez-Paz N, Chng WJ, McClure RF, Blood E, Oken MM, Van Ness B, James CD, Kurtin PJ, Henderson K, Ahmann GJ, Gertz M, Lacy M, Dispenzieri A, Greipp PR, Fonseca R |title=Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications |journal=Blood |volume=109 |issue=3 |pages=1228–32 |date=February 2007 |pmid=16840723 |doi=10.1182/blood-2006-05-024661 |url=}}</ref><ref name="pmid15603511">{{cite journal |vauthors=Seemann S, Maurici D, Olivier M, Caron de Fromentel C, Hainaut P |title=The tumor suppressor gene TP53: implications for cancer management and therapy |journal=Crit Rev Clin Lab Sci |volume=41 |issue=5-6 |pages=551–83 |date=2004 |pmid=15603511 |doi=10.1080/10408360490504952 |url=}}</ref> | |||
* The loss of [[Tumor suppressor|tumor suppressors]] allows the [[Cell (biology)|cells]] to survive and grow without [[Cell cycle checkpoint|check points]]. | |||
{| class="wikitable" style="width:40%; height:100px" border="1" | |||
|+ | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Tumor suppressor genes commonly affected in myeloma | |||
|- | |||
|''[[FAM46C]] (''family with sequence similarity 46, member C) | |||
|- | |||
|''[[DIS3]] (''Exosome complex exonuclease RRP44) | |||
|- | |||
|''[[CYLD (gene)|CYLD]] (''[[Cylindromatosis]]) | |||
|- | |||
|Baculoviral IAP repeat containing protein 2 (''BIRC2''; also known as cIAP1) | |||
|- | |||
|''[[BIRC3 gene|BIRC3]] ([[BIRC3 gene|Baculoviral IAP repeat containing protein 3]])'' | |||
|- | |||
|[[TRAF3|tumor necrosis factor receptor associated factor 3]] (''[[TRAF3]]'') | |||
|- | |||
|[[CDKN2C]] | |||
|- | |||
|[[CDKN2A]] | |||
|- | |||
|''[[TP53]]'' | |||
|} | |||
===== '''''NFKB alterations''''' ===== | |||
* '''[[NF-κB]]''' (nuclear factor kappa-light-chain-enhancer of activated B cells) is a [[protein]] complex which regulates [[DNA]] [[Transcription (genetics)|transcription]], production of [[Cytokines|cytokine]] and cell survival. [[NF-κB]] plays a key role in generating cellular responses to stimuli such as stress, [[free radicals]], [[heavy metals]], ultraviolet radiations, and foreign [[antigens]]. [[NF-κB]] is also crucial in response of the [[Immune systems|immune system]] towards [[infections]].<ref name="pmid17072321">{{cite journal |vauthors=Gilmore TD |title=Introduction to NF-kappaB: players, pathways, perspectives |journal=Oncogene |volume=25 |issue=51 |pages=6680–4 |date=October 2006 |pmid=17072321 |doi=10.1038/sj.onc.1209954 |url=}}</ref><ref name="pmid17183360">{{cite journal |vauthors=Perkins ND |title=Integrating cell-signalling pathways with NF-kappaB and IKK function |journal=Nat. Rev. Mol. Cell Biol. |volume=8 |issue=1 |pages=49–62 |date=January 2007 |pmid=17183360 |doi=10.1038/nrm2083 |url=}}</ref><ref name="pmid12795416">{{cite journal |vauthors=Tian B, Brasier AR |title=Identification of a nuclear factor kappa B-dependent gene network |journal=Recent Prog. Horm. Res. |volume=58 |issue= |pages=95–130 |date=2003 |pmid=12795416 |doi= |url=}}</ref> | |||
* Deregulated '''[[NF-κB]]''' system leads to increased [[growth factors]] and [[cytokines]] such as [[IL-6]] which promote growth and survival in [[Multiple myeloma|myeloma]] cells.<ref name="pmid12795416">{{cite journal |vauthors=Tian B, Brasier AR |title=Identification of a nuclear factor kappa B-dependent gene network |journal=Recent Prog. Horm. Res. |volume=58 |issue= |pages=95–130 |date=2003 |pmid=12795416 |doi= |url=}}</ref> | |||
* [[Mutations]] in multiple [[genes]] can cause activation of [[NF-κB]] system which in turn plays a role in [[pathogenesis]] of [[multiple myeloma]]. Some of these [[mutations]], that have been documented in [[genes]] regulating [[NF-κB]] pathway, are mentioned below.<ref name="pmid29772694">{{cite journal |vauthors=Roy P, Sarkar UA, Basak S |title=The NF-κB Activating Pathways in Multiple Myeloma |journal=Biomedicines |volume=6 |issue=2 |pages= |date=May 2018 |pmid=29772694 |pmc=6027071 |doi=10.3390/biomedicines6020059 |url=}}</ref><ref name="pmid21430775">{{cite journal |vauthors=Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR |title=Initial genome sequencing and analysis of multiple myeloma |journal=Nature |volume=471 |issue=7339 |pages=467–72 |date=March 2011 |pmid=21430775 |pmc=3560292 |doi=10.1038/nature09837 |url=}}</ref> | |||
{| | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Genes mutated associated with canonical signaling | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Genes mutated associated with non-canonical signaling | |||
|- | |||
| align="left" style="background:#F5F5F5;" + |''[[TLR4]]'' | |||
''TNFRSF1A'' | |||
''IKBKB'' | |||
''IKBIP'' | |||
''[[CARD11]]'' | |||
''[[MAP3K1]]'' | |||
''[[RIPK4]]'' | |||
''[[CYLD (gene)|CYLD]]'' | |||
''[[BTRC (gene)|BTRC]]'' | |||
| align="left" style="background:#F5F5F5;" + |''[[MAP3K14]]'' | |||
''[[TRAF3]]'' | |||
|} | |||
===== Epigenetic changes in multiple myeloma ===== | |||
* [[Epigenetics|Epigenetic]] events such as [[methylation]] of [[DNA]] and [[histones]] play an important role in [[pathogenesis]] of [[multiple myeloma]]. | |||
* Of these events, the most notable is the global [[DNA methylation|hypomethylation]] and [[gene]]-specific [[DNA methylation|hypermethylation]] that takes place during progression of [[Monoclonal gammopathy of undetermined significance|MGUS]] to [[Multiple myeloma|myeloma]].<ref name="pmid24688108" /> | |||
* A subset of [[patients]] with t(4;14) [[translocation]] show prominent [[DNA methylation]] changes that lead to MMSET overexpression.<ref name="pmid24688108" /><ref name="pmid9207791">{{cite journal |vauthors=Chesi M, Nardini E, Brents LA, Schröck E, Ried T, Kuehl WM, Bergsagel PL |title=Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3 |journal=Nat. Genet. |volume=16 |issue=3 |pages=260–4 |date=July 1997 |pmid=9207791 |pmc=3901950 |doi=10.1038/ng0797-260 |url=}}</ref> | |||
* Other dysregulations related to [[epigenetics]] in [[multiple myeloma]] may effect KDM6A ([[UTX (gene)|UTX]]), a [[histone]] [[demethylase]], mixed lineage leukemia ([[MLL]]) protein, [[KDM6B|lysine demethylase 6B]] ([[KDM6B]]) and [[HOXA9|homeobox A9]] ([[HOXA9]]).<ref name="pmid24688108" /><ref name="pmid19330029">{{cite journal |vauthors=van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA |title=Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer |journal=Nat. Genet. |volume=41 |issue=5 |pages=521–3 |date=May 2009 |pmid=19330029 |pmc=2873835 |doi=10.1038/ng.349 |url=}}</ref> | |||
===== Bone marrow microenvironment and multiple myeloma ===== | |||
* [[Bone marrow]] microenvironment comprises of [[Bone marrow|bone marrow stromal cells]] (BMSCs), [[Extracellular matrix|extracellular matrix (ECM) proteins]] such as [[collagen]], [[fibronectin]] and [[laminin]], and the [[extracellular fluid]] containing [[cytokines]] and [[growth factors]]. This microenvironment is conducive to normal [[hematopoiesis]] but this also helps [[Multiple myeloma|myeloma]] cells to have increased [[replication]] activity and anti-[[Apoptosis|apoptotic]] resistance.<ref name="pmid23176722" /><ref name="pmid24688108" /><ref name="pmid16765041">{{cite journal |vauthors=Mitsiades CS, Mitsiades NS, Munshi NC, Richardson PG, Anderson KC |title=The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions |journal=Eur. J. Cancer |volume=42 |issue=11 |pages=1564–73 |date=July 2006 |pmid=16765041 |doi=10.1016/j.ejca.2005.12.025 |url=}}</ref> | |||
* One of the key processes is the interaction and adherence of [[Multiple myeloma|myeloma]] cells with [[Bone marrow|bone marrow stromal cells]] (BMSCs) and [[Extracellular matrix|extracellular matrix proteins]] through [[cellular adhesion molecules]] (CAMs) such as [[CD44]] (H-CAM), [[CD56]] (N-CAM), members of the [[CD49a|CD49]] [[Integrins|integrin]] family (including very-late antigen VLA-4 and VLA-5), lymphocyte function-associated antigen-1, [[Syndecan 1|syndecan]]-1, and [[Selectins|selectin]].<ref name="pmid23176722" /><ref name="pmid24688108" /><ref name="pmid9081202">{{cite journal |vauthors=Teoh G, Anderson KC |title=Interaction of tumor and host cells with adhesion and extracellular matrix molecules in the development of multiple myeloma |journal=Hematol. Oncol. Clin. North Am. |volume=11 |issue=1 |pages=27–42 |date=February 1997 |pmid=9081202 |doi= |url=}}</ref> | |||
* This interaction leads to activation of p42/44 [[mitogen-activated protein kinase]], and [[NF-κB|nuclear factor κB]] ([[NF-κB]]) which cause increased adhesion molecules, both on [[Multiple myeloma|myeloma]] cells and [[Bone marrow|bone marrow stromal cells]]. These events ultimately lead to increased production of [[cytokines]]. Of these cytokines [[IL-6]], [[TNFα]], [[BAFF receptor|BAFF]], [[IGF]] and [[HGFAC|HGF]] are of particular importance.<ref name="pmid23176722" /><ref name="pmid24688108" /><ref name="pmid8562936">{{cite journal |vauthors=Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA, Anderson KC |title=Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B |journal=Blood |volume=87 |issue=3 |pages=1104–12 |date=February 1996 |pmid=8562936 |doi= |url=}}</ref><ref name="pmid15070697">{{cite journal |vauthors=Moreaux J, Legouffe E, Jourdan E, Quittet P, Rème T, Lugagne C, Moine P, Rossi JF, Klein B, Tarte K |title=BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone |journal=Blood |volume=103 |issue=8 |pages=3148–57 |date=April 2004 |pmid=15070697 |pmc=2387243 |doi=10.1182/blood-2003-06-1984 |url=}}</ref><ref name="pmid11830493">{{cite journal |vauthors=Derksen PW, Keehnen RM, Evers LM, van Oers MH, Spaargaren M, Pals ST |title=Cell surface proteoglycan syndecan-1 mediates hepatocyte growth factor binding and promotes Met signaling in multiple myeloma |journal=Blood |volume=99 |issue=4 |pages=1405–10 |date=February 2002 |pmid=11830493 |doi= |url=}}</ref> | |||
* These events cause localization of [[Tumor cell|tumor cells]] in [[bone marrow]], increased proliferation and survival, and [[resistance]] to [[apoptosis]] and [[chemotherapy]]. [[Bone marrow]] niche thus plays a crucial role in [[pathogenesis]] of [[multiple myeloma]].<ref name="pmid23176722" /><ref name="pmid24688108" /><ref name="pmid8562936" /><ref name="pmid15070697" /><ref name="pmid11830493" /> | |||
* Decreased [[cellular adhesion molecules]] such as [[CD56]] and [[Chemokines|chemokine]] receptors such as [[CXCR4]] on [[Multiple myeloma|myeloma]] cells lead to decreased adhesion to [[Bone marrow|bone marrow stromal cells]] and [[Immune system|immune]] evasion, allowing the [[Multiple myeloma|myeloma]] cells to spread outside the [[bone marrow]].<ref name="pmid9844928">{{cite journal |vauthors=Pellat-Deceunynck C, Barillé S, Jego G, Puthier D, Robillard N, Pineau D, Rapp MJ, Harousseau JL, Amiot M, Bataille R |title=The absence of CD56 (NCAM) on malignant plasma cells is a hallmark of plasma cell leukemia and of a special subset of multiple myeloma |journal=Leukemia |volume=12 |issue=12 |pages=1977–82 |date=December 1998 |pmid=9844928 |doi= |url=}}</ref><ref name="pmid1382543">{{cite journal |vauthors=Barker HF, Hamilton MS, Ball J, Drew M, Franklin IM |title=Expression of adhesion molecules LFA-3 and N-CAM on normal and malignant human plasma cells |journal=Br. J. Haematol. |volume=81 |issue=3 |pages=331–5 |date=July 1992 |pmid=1382543 |doi= |url=}}</ref> | |||
===== Cytokines in multiple myeloma pathogenesis ===== | |||
* A variety of cytokines have been implicated in [[pathogenesis]] of [[multiple myeloma]]. Some of the relevant [[cytokines]], their mechanisms and effects on [[Tumor|tumor cells]] have been summarized in the table below.<ref name="pmid23176722" /><ref name="pmid12965277">{{cite journal |vauthors=Seidl S, Kaufmann H, Drach J |title=New insights into the pathophysiology of multiple myeloma |journal=Lancet Oncol. |volume=4 |issue=9 |pages=557–64 |date=September 2003 |pmid=12965277 |doi= |url=}}</ref><ref name="pmid11753617">{{cite journal |vauthors=Gupta D, Treon SP, Shima Y, Hideshima T, Podar K, Tai YT, Lin B, Lentzsch S, Davies FE, Chauhan D, Schlossman RL, Richardson P, Ralph P, Wu L, Payvandi F, Muller G, Stirling DI, Anderson KC |title=Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications |journal=Leukemia |volume=15 |issue=12 |pages=1950–61 |date=December 2001 |pmid=11753617 |doi= |url=}}</ref><ref name="pmid11753617">{{cite journal |vauthors=Gupta D, Treon SP, Shima Y, Hideshima T, Podar K, Tai YT, Lin B, Lentzsch S, Davies FE, Chauhan D, Schlossman RL, Richardson P, Ralph P, Wu L, Payvandi F, Muller G, Stirling DI, Anderson KC |title=Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications |journal=Leukemia |volume=15 |issue=12 |pages=1950–61 |date=December 2001 |pmid=11753617 |doi= |url=}}</ref><ref name="pmid8978296">{{cite journal |vauthors=Chauhan D, Kharbanda S, Ogata A, Urashima M, Teoh G, Robertson M, Kufe DW, Anderson KC |title=Interleukin-6 inhibits Fas-induced apoptosis and stress-activated protein kinase activation in multiple myeloma cells |journal=Blood |volume=89 |issue=1 |pages=227–34 |date=January 1997 |pmid=8978296 |doi= |url=}}</ref><ref name="pmid11494147">{{cite journal |vauthors=Hideshima T, Chauhan D, Schlossman R, Richardson P, Anderson KC |title=The role of tumor necrosis factor alpha in the pathophysiology of human multiple myeloma: therapeutic applications |journal=Oncogene |volume=20 |issue=33 |pages=4519–27 |date=July 2001 |pmid=11494147 |doi=10.1038/sj.onc.1204623 |url=}}</ref><ref name="pmid10210775">{{cite journal |vauthors=Jourdan M, Tarte K, Legouffe E, Brochier J, Rossi JF, Klein B |title=Tumor necrosis factor is a survival and proliferation factor for human myeloma cells |journal=Eur. Cytokine Netw. |volume=10 |issue=1 |pages=65–70 |date=March 1999 |pmid=10210775 |pmc=2025696 |doi= |url=}}</ref><ref name="pmid16818641">{{cite journal |vauthors=Tai YT, Li XF, Breitkreutz I, Song W, Neri P, Catley L, Podar K, Hideshima T, Chauhan D, Raje N, Schlossman R, Richardson P, Munshi NC, Anderson KC |title=Role of B-cell-activating factor in adhesion and growth of human multiple myeloma cells in the bone marrow microenvironment |journal=Cancer Res. |volume=66 |issue=13 |pages=6675–82 |date=July 2006 |pmid=16818641 |doi=10.1158/0008-5472.CAN-06-0190 |url=}}</ref><ref name="pmid8822946">{{cite journal |vauthors=Georgii-Hemming P, Wiklund HJ, Ljunggren O, Nilsson K |title=Insulin-like growth factor I is a growth and survival factor in human multiple myeloma cell lines |journal=Blood |volume=88 |issue=6 |pages=2250–8 |date=September 1996 |pmid=8822946 |doi= |url=}}</ref><ref name="pmid10753844">{{cite journal |vauthors=Dankbar B, Padró T, Leo R, Feldmann B, Kropff M, Mesters RM, Serve H, Berdel WE, Kienast J |title=Vascular endothelial growth factor and interleukin-6 in paracrine tumor-stromal cell interactions in multiple myeloma |journal=Blood |volume=95 |issue=8 |pages=2630–6 |date=April 2000 |pmid=10753844 |doi= |url=}}</ref><ref name="pmid20395418">{{cite journal |vauthors=Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, Pai C, Amin S, Tai YT, Richardson PG, Ghobrial IM, Treon SP, Daley JF, Anderson KC, Kutok JL, Munshi NC |title=Elevated IL-17 produced by TH17 cells promotes myeloma cell growth and inhibits immune function in multiple myeloma |journal=Blood |volume=115 |issue=26 |pages=5385–92 |date=July 2010 |pmid=20395418 |pmc=2902136 |doi=10.1182/blood-2009-10-246660 |url=}}</ref><ref name="pmid20664052">{{cite journal |vauthors=Noonan K, Marchionni L, Anderson J, Pardoll D, Roodman GD, Borrello I |title=A novel role of IL-17-producing lymphocytes in mediating lytic bone disease in multiple myeloma |journal=Blood |volume=116 |issue=18 |pages=3554–63 |date=November 2010 |pmid=20664052 |pmc=4017298 |doi=10.1182/blood-2010-05-283895 |url=}}</ref> | |||
{| class="wikitable" | |||
|+ | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Cytokines | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Mechanism | |||
! style="background:#4479BA; color: #FFFFFF;" align="center" + |Effects on tumor cells and pathogenesis | |||
|- | |||
|[[Interleukin 6]] | |||
[[IL-6]] | |||
|Activates [[Signal transduction pathway|signal transduction pathways]] | |||
([[JAK-STAT signaling pathway|JAK/STAT3]] and [[PI3K]]/[[Akt]]) | |||
| | |||
* Increased proliferation | |||
* Increased production of [[vascular endothelial growth factor]] ([[VEGF]]) | |||
* Decreased [[apoptosis]] | |||
* [[Drug resistance]] | |||
* Increased [[osteoclasts]] [[differentiation]] | |||
|- | |||
|[[Tumor necrosis factor|Tumor necrosis factor α]] | |||
[[TNF-α|TNF]]-α | |||
|Activation of [[NF-κB]] | |||
Activation of the [[MAPK]] pathways | |||
| | |||
* Increased survival | |||
* Increased proliferation | |||
* Increased [[cell migration]] | |||
* Increased [[Inflammation|inflammatory]] mediators | |||
* Increased [[Osteoclasts|osteoclast]] [[differentiation]] | |||
* Increased [[bone]] resorption | |||
|- | |||
|[[B-cell activating factor]] | |||
([[B-cell activating factor|BAFF]]) | |||
| [[ | |Activation of [[NF-κB]] | ||
| [[ | | | ||
* Increased survival | |||
* Increased [[adhesion]] | |||
|- | |||
|[[Insulin-like growth factor-1]] | |||
([[IGF-1]]) | |||
|Activation of [[PI3K]]/[[Akt]] | |||
Activation of [[IKK2|IKK]]/[[NF-κB]] | |||
| | |||
* Increased growth and proliferation | |||
* Decreased apoptosis and increased survival | |||
|- | |||
|[[Vascular endothelial growth factor]] | |||
([[VEGF]]) | |||
|[[VEGF receptors|VEGF Receptor]] activation | |||
| | |||
* Increased [[angiogenesis]] | |||
* Increased [[tumor]] cell migration | |||
* Increased [[growth]], | |||
* Increased survival | |||
* Increased [[IL-6]] production | |||
|- | |||
|[[Interleukin 17]] | |||
([[Interleukin 17|IL-17]]) | |||
|[[Interleukin 17]] receptors activation | |||
| | |||
* Increased survival | |||
* Increased [[cytokines]] production | |||
* Lytic [[bone]] lesions | |||
|} | |} | ||
===Microscopic Pathology=== | ===Pathophysiology of renal involvement=== | ||
Abnormal [[antibody]] fragments are produced in multiple myeloma and are deposited in various organs, such as the kidneys. There are three major forms of renal damage in patients with multiple myeloma. | |||
*'''Cast [[nephropathy]]''': End-organ damage to the kidneys is typically due to light chain cast [[nephropathy]]. The pathophysiology of this type of renal involvement is based on light chain deposition in the [[renal tubules]], which results in obstruction. Free light chains are readily filtered at the [[glomerulus]] and are reabsorbed in the proximal tubule of the [[nephron]]. This reabsorption occurs via the megalin-cubulin transport system.<ref name="pmid27526708">{{cite journal| author=Finkel KW, Cohen EP, Shirali A, Abudayyeh A, American Society of Nephrology Onco-Nephrology Forum| title=Paraprotein-Related Kidney Disease: Evaluation and Treatment of Myeloma Cast Nephropathy. | journal=Clin J Am Soc Nephrol | year= 2016 | volume= 11 | issue= 12 | pages= 2273-2279 | pmid=27526708 | doi=10.2215/CJN.01640216 | pmc=5142056 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27526708 }} </ref> In patients with multiple myeloma, there is excess production of free [[Light chain|light chains]], and the ability of the nephron to resorb light chains in the [[proximal tubule]] cannot meet the demands of the freely filtered [[Light chain|light chains]]. This results in excess secretion of free [[Light chain|light chains]] in the urine (known as Bence-Jones protein). [[Eosinophilic]] proteinaceous casts and crystalline structures can be seen. Cast formation occurs in the tubules due to excess abundance of free light chains that interact with [[Tamm-Horsfall protein|Tamm-Horsfall proteins]] in the thick ascending [[loop of Henle]].<ref name="pmid27526708">{{cite journal| author=Finkel KW, Cohen EP, Shirali A, Abudayyeh A, American Society of Nephrology Onco-Nephrology Forum| title=Paraprotein-Related Kidney Disease: Evaluation and Treatment of Myeloma Cast Nephropathy. | journal=Clin J Am Soc Nephrol | year= 2016 | volume= 11 | issue= 12 | pages= 2273-2279 | pmid=27526708 | doi=10.2215/CJN.01640216 | pmc=5142056 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27526708 }} </ref> Tubular obstruction ensues, triggering local inflammation which results in [[interstitial nephritis]] and fibrosis.<ref name="pmid27526708">{{cite journal| author=Finkel KW, Cohen EP, Shirali A, Abudayyeh A, American Society of Nephrology Onco-Nephrology Forum| title=Paraprotein-Related Kidney Disease: Evaluation and Treatment of Myeloma Cast Nephropathy. | journal=Clin J Am Soc Nephrol | year= 2016 | volume= 11 | issue= 12 | pages= 2273-2279 | pmid=27526708 | doi=10.2215/CJN.01640216 | pmc=5142056 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=27526708 }} </ref> The onset of cast nephropathy can be very quick, requiring prompt treatment. Risk factors for development of [[Nephropathy|cast nephropathy]] include monoclonal immunoglobulin secretion of >10 g/day, sepsis, and volume depletion.<ref name="pmid23868898">{{cite journal| author=Heher EC, Rennke HG, Laubach JP, Richardson PG| title=Kidney disease and multiple myeloma. | journal=Clin J Am Soc Nephrol | year= 2013 | volume= 8 | issue= 11 | pages= 2007-17 | pmid=23868898 | doi=10.2215/CJN.12231212 | pmc=3817918 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=23868898 }} </ref> Patients can also develop [[Fanconi syndrome]], resulting in dysfunctional reabsorption ability by the [[proximal tubule]], and type II [[renal tubular acidosis]]. | |||
*'''Monoclonal immunoglobulin deposition disease (MIDD)''': In this subtype of renal involvement by multiple myeloma, the initial pathophysiological process is filtration of monoclonal immunoglobulins and subsequent deposition of [[immunoglobulins]] along the tubular or glomerular basement membrane.<ref name="pmid23868898">{{cite journal| author=Heher EC, Rennke HG, Laubach JP, Richardson PG| title=Kidney disease and multiple myeloma. | journal=Clin J Am Soc Nephrol | year= 2013 | volume= 8 | issue= 11 | pages= 2007-17 | pmid=23868898 | doi=10.2215/CJN.12231212 | pmc=3817918 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=23868898 }} </ref> Deposits of immunoglobulin can have a similar appearance as [[Kimmelstein-Wilson Disease|Kimmelstein-Wilson lesions]] (seen in diabetes). The immunoglobulins can appear fibroblast-like. | |||
*'''Light chain [[amyloidosis]]''': The pathophysiology of renal involvement by light chain amyloidosis begins with ''beta''-pleated sheet formation in the [[tubules]] or glomeruli. ''Beta''-pleated sheets form as a result of electrostatic interactions between [[heparan sulfate]] proteoglycan and amyloid proteins. Amyloid fibrils usually consist of immunoglobulin light chains (usually ''lambda'' light chain) and deposit in the [[basement membrane]]. The size of the fibrils vary from 7 to 10 nanometers. A diagnosis of this type of renal involvement is made by the visualization of apple green birefringence upon [[Congo red|Congo red staining]] of the renal specimen.<ref name="pmid23868898">{{cite journal| author=Heher EC, Rennke HG, Laubach JP, Richardson PG| title=Kidney disease and multiple myeloma. | journal=Clin J Am Soc Nephrol | year= 2013 | volume= 8 | issue= 11 | pages= 2007-17 | pmid=23868898 | doi=10.2215/CJN.12231212 | pmc=3817918 | url=https://www.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&tool=sumsearch.org/cite&retmode=ref&cmd=prlinks&id=23868898 }} </ref> It is frequently associated with nephrotic range [[proteinuria]], in which greater than 3 grams of protein is excreted daily. | |||
===Pathophysiology of osseous involvement=== | |||
[[Bone]] [[disease]] characterized by progressive osteolytic [[bone]] lesions leading to [[bone resorption]] is hallmark of [[multiple myeloma]]. Abnormal [[bone]] remodeling is thought to be the cause. It has been reported that up to 80% of the [[patients]] have characteristic osteolytic bone lesions at presentation and 60% of the patients with [[multiple myeloma]] will develop at least one pathological [[Bone fracture|fracture]] at some stage.The [[pathophysiology]] of [[Bone|bony]] involvement in [[multiple myeloma]] is complex and is briefly described here..<ref name="pmid29330358">{{cite journal |vauthors=Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA |title=Pathogenesis of bone disease in multiple myeloma: from bench to bedside |journal=Blood Cancer J |volume=8 |issue=1 |pages=7 |date=January 2018 |pmid=29330358 |pmc=5802524 |doi=10.1038/s41408-017-0037-4 |url=}}</ref><ref name="pmid23690408">{{cite journal |vauthors=Terpos E, Morgan G, Dimopoulos MA, Drake MT, Lentzsch S, Raje N, Sezer O, García-Sanz R, Shimizu K, Turesson I, Reiman T, Jurczyszyn A, Merlini G, Spencer A, Leleu X, Cavo M, Munshi N, Rajkumar SV, Durie BG, Roodman GD |title=International Myeloma Working Group recommendations for the treatment of multiple myeloma-related bone disease |journal=J. Clin. Oncol. |volume=31 |issue=18 |pages=2347–57 |date=June 2013 |pmid=23690408 |pmc=4878084 |doi=10.1200/JCO.2012.47.7901 |url=}}</ref><ref name="pmid20376081">{{cite journal |vauthors=Terpos E, Berenson J, Cook RJ, Lipton A, Coleman RE |title=Prognostic variables for survival and skeletal complications in patients with multiple myeloma osteolytic bone disease |journal=Leukemia |volume=24 |issue=5 |pages=1043–9 |date=May 2010 |pmid=20376081 |doi=10.1038/leu.2010.62 |url=}}</ref> | |||
===== Increased osteoclastic activity ===== | |||
[[Osteoclasts]] are large, multinucleated cells of [[Monocytes|monocyte]]–[[macrophage]] lineage and play a crucial role in [[bone]] remodeling. [[Multiple myeloma|Myeloma]] cells, in addition to their direct interaction with other [[cells]], produce and release a number of factors which promote [[Osteoclasts|osteoclast]] differentiation and activation. Some of these factors and interactions have been described below in the tables.<ref name="pmid29330358">{{cite journal |vauthors=Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA |title=Pathogenesis of bone disease in multiple myeloma: from bench to bedside |journal=Blood Cancer J |volume=8 |issue=1 |pages=7 |date=January 2018 |pmid=29330358 |pmc=5802524 |doi=10.1038/s41408-017-0037-4 |url=}}</ref><ref name="pmid18406675">{{cite journal |vauthors=Edwards CM, Zhuang J, Mundy GR |title=The pathogenesis of the bone disease of multiple myeloma |journal=Bone |volume=42 |issue=6 |pages=1007–13 |date=June 2008 |pmid=18406675 |pmc=2474770 |doi=10.1016/j.bone.2008.01.027 |url=}}</ref><ref name="pmid25187738">{{cite journal |vauthors=Hameed A, Brady JJ, Dowling P, Clynes M, O'Gorman P |title=Bone disease in multiple myeloma: pathophysiology and management |journal=Cancer Growth Metastasis |volume=7 |issue= |pages=33–42 |date=2014 |pmid=25187738 |pmc=4133035 |doi=10.4137/CGM.S16817 |url=}}</ref> | |||
{| class="wikitable" | |||
|+ | |||
!style="background:#4479BA; color: #FFFFFF;" align="center" + |Cell-cell interactions | |||
!style="background:#4479BA; color: #FFFFFF;" align="center" + |Consequences | |||
|- | |||
|'''[[Multiple myeloma|Myeloma cells]] to [[bone marrow]] stromal [[cells]]''' | |||
| | |||
* Decreased production of [[OPGs|OPG]] | |||
* Increased [[RANKL]] expression | |||
* Activation of the [[NF-kB]] system that has following effects on [[Multiple myeloma|myeloma cells]]: | |||
** ↑ growth | |||
** ↑ survival | |||
** ↑ [[drug resistance]] | |||
** migration | |||
|- | |||
|'''[[Integrins|Alpha4-beta1 integrin]] to [[VCAM-1|vascular cell adhesion molecule 1]] ([[VCAM-1|VCAM-1)]] interaction''' | |||
| | |||
* Increased [[Osteoclast|osteoclastic]] activity independent of [[IL-6]], [[TNF]] or [[Parathyroid hormone-related protein|PTHrP]]. | |||
* Increased [[RANKL]] expression | |||
|- | |||
|'''[[Multiple myeloma|Myeloma cells]] to [[osteoblasts]]''' | |||
|Decreased production of [[OPGs|OPG]] | |||
|- | |||
|'''[[Multiple myeloma|Myeloma cells]] to [[osteoclasts]]''' | |||
|Direct adherence of [[Multiple myeloma|myeloma cells]] to [[osteoclasts]] may result in | |||
* [[Multiple myeloma|myeloma cell]] proliferation | |||
* ↑ [[Osteoclasts|osteoclastic]] [[differentiation]] | |||
* ↑ [[Angiogenic|proangiogenic]] factors | |||
|- | |||
|'''[[Multiple myeloma|Myeloma cells]] to [[immune cells]]''' | |||
|Increased production of [[cytokines]], [[chemokines]] and factors associated with growth, | |||
survival and migration. | |||
|} | |||
. | |||
{| class="wikitable" | |||
|+ | |||
!Molecular pathways and factors associated with increased osteoclastic activity | |||
!Association with multiple myeloma | |||
|- | |||
|'''RANK/RANKL pathway''' | |||
'''Binding of [[RANKL]] to [[RANK]] → fusion of [[Osteoclasts|osteoclast]] precursors into multinucleated cells → mature [[Osteoclast|osteoclasts]] → ↑ [[bone resorption]]''' | |||
'''[[Osteoprotegerin]] ([[OPGs|OPG]]) is a soluble decoy [[receptor]] for [[RANKL]] → ↓ binding of [[RANKL]] to [[RANK]] → ↓ [[bone resorption]]''' | |||
* [[RANK]] is a [[transmembrane receptor]] of [[TNF]] superfamily and is expressed by [[osteoclast]] precursors | |||
* [[RANKL]], a [[membrane]]-bound [[protein]] on BMSCs of [[Osteoblasts|osteoblastic]] lineage and activated [[T-lymphocytes]], is a [[Cytokines|cytokine]] | |||
* [[Osteoprotegerin]] ([[OPGs|OPG]]), released by BMSCs and [[osteoblasts]], is a member of the [[TNF]] superfamily | |||
* '''<big>↑</big>''' [[RANKL]] and/or '''<big>↓</big>''' [[OPGs|OPG]] '''<big>→</big>''' '''<big>↑</big>''' [[Bone resorption]] and destruction | |||
|[[Multiple myeloma|Myeloma]] cells lead to increased [[RANKL]] and decreased [[OPGs|OPG]] causing [[bone resorption]] and destruction. [[RANKL]]/[[OPGs|OPG]] ratio is an idependent | |||
[[prognostic]] factor in multiple myeloma | |||
* [[Multiple myeloma|Myeloma]] cells produce and release soluble [[RANKL]] → '''<big>↑</big>''' [[Bone resorption]] | |||
* [[Plasma cells]] secrete [[Parathyroid hormone-related protein|PTHrP]] that stimulates [[Paracrine signalling|paracrine pathway]] → ↑ expression of [[RANKL]] by [[osteoblasts]] and BMSCs → '''<big>↑</big>''' [[Bone resorption]] | |||
* [[Syndecan 1|Syndecan-1]], an heparan sulfate proteoglycan expressed by [[Multiple myeloma|myeloma]] cells, binds to [[OPG]] → [[endocytosis]] and degradation of [[OPG]] by [[Multiple myeloma|myeloma]] cells → '''<big>↑</big>''' [[Bone resorption]] | |||
|- | |||
|'''[[Notch signaling pathway|Notch pathway]]''' | |||
'''Binding of [[Notch family of receptors|Notch receptors]] to its [[ligands]] → ↑ [[RANKL]] production → ↑ [[bone resorption]]''' | |||
* [[Notch family of receptors|Notch family]] contains four [[transmembrane receptors]] ([[Notch family of receptors|Notch]] 1–4) | |||
* Their '''[[ligands]]''' include Jagged 1,2 and [[Delta-like 1|Delta-like]] 1,3,4 | |||
|[[Multiple myeloma|Myeloma]] cells express [[Notch family of receptors|Notch]] 1,2,3 and their '''[[ligands]]''' Jagged 1,2 and [[Delta-like 1|Delta-like]] 1,3,4. This pathway, in addition to [[bone resorption]], may also be involved in [[metastasis]] of [[Multiple myeloma|myeloma]] cells by increasing expression of [[Cell adhesion molecule|adhesion molecules]], migratory [[chemokines]], and angiogenetic factors, and disruption of the immune surveillance. | |||
* [[Notch family of receptors|Notch receptors]] on [[Multiple myeloma|myeloma]]<nowiki/>cells can bind to Jagged 1,2 '''[[ligands]]''' on the same [[cell]] (homotypical interaction) → ↑ [[RANKL]] | |||
* [[Notch family of receptors|Notch receptors]] on [[Multiple myeloma|myeloma]] cells can bind to Jagged or Delta-like '''[[ligands]]''' on adjacent BMSCs and [[Multiple myeloma|myeloma]] cells (heterotypical interaction) → ↑ [[RANKL]] | |||
* [[Notch family of receptors|Notch receptors]] on BMSCs can bind to Jagged '''[[ligands]]''' on [[Multiple myeloma|myeloma]] cells → ↑ [[RANKL]] | |||
|- | |||
|'''[[Macrophage inflammatory protein|CCL-3 (MIP-1α)]]/CCL-20''' | |||
* [[Macrophage inflammatory protein|Chemokine (C-C motif) ligand 3 (CCL-3)]], previously known as [[Macrophage inflammatory protein|macrophage inflammatory protein-1α (]]MIP-1α), is a [[chemokine]] that binds to [[CCR1]] and [[CCR5 receptor|CCR5]]. | |||
* Chemokine (C-C motif) ligand 20 (CCL-20) is a [[Chemokines|chemokine]] involved in [[Th17]] pathway that binds to CCR6 and has been implicated in [[Osteoclasts|osteoclastogenesis]] and osteolytic lesions. | |||
* [[Macrophage inflammatory protein|CCL-3]] may: | |||
** attract [[Osteoclasts|osteoclast]] precursors and induce [[Osteoclasts|osteoclastogenesis]] | |||
** potentiate [[RANKL]] and [[Interleukin 6|IL-6]] effects on [[osteoclasts]] | |||
** down-regulate [[RUNX2]] and osterix → ↓ [[osteoblast]] activity | |||
** ↓ bone mineralization | |||
** promote survival of [[Multiple myeloma|myeloma]] cells | |||
** promote migration of [[Multiple myeloma|myeloma]] cells | |||
| | |||
* [[Multiple myeloma]] patients have high levels of [[Macrophage inflammatory protein|CCL-3]]/CCL-20 detected in their [[bone marrow]] and [[serum]]. | |||
* In [[Multiple myeloma|myeloma cells]] with [[Translocations|translocation]] t(4;14), up-regulation of [[FGFR3|fibroblast growth factor 3]] ([[FGFR3|FGFR]]-3) leads to induction of [[Macrophage inflammatory protein|CCL-3.]] | |||
* High levels of [[Macrophage inflammatory protein|CCL-3]]/CCL-20 in [[Multiple myeloma|myeloma]] patients correlate positively with [[Bone cell|bone]] disease and negatively with survival. | |||
|- | |||
|'''[[Activin|Activin A]]''' | |||
Binds to type II transmembrane serine/threonine kinase receptor (ActRIIA/B) → recruitment and phosphorylation the type I receptor | |||
(ActRI, also called activin receptor-like kinase 4 (ALK4) receptor) → [[heterodimer]] formation → activation of the [[Smad]] signaling cascade → translocation of [[Smad2]]/[[Smad3|3]]/[[Smad4|4]] complex in the [[nucleus]] → action as [[Transcription factor|transcriptional factor]] → [[RANK]] expression and activation of [[NF-κB|NF-κB pathway]] → increased [[osteoclast]] [[differentiation]] | |||
* It belongs to [[TGFβ]] superfamily. | |||
* May also be involved in [[Akt]]/[[PI3K]], [[MAPK]]/[[ERK]], [[JNK]], and [[WNT1|WNT]]/[[Beta-catenin|β-catenin pathways]]. | |||
| | |||
* [[Multiple myeloma]] patients have high levels of [[activin]] A detected in their [[bone marrow]] and [[serum]]. | |||
* Increased [[activin]] A levels may be associated with advanced [[bone]] disease and worse [[prognosis]] in [[multiple myeloma]]. | |||
* It has been suggested that interaction between BMSCs and [[Multiple myeloma|myeloma cells]] may induce secretion of [[activin]] A. | |||
* It is also involved in inhibition of [[BMP]] signaling in [[Multiple myeloma|myeloma cells]]. | |||
|- | |||
|'''Osteopontin''' | |||
* a non-collagenous [[bone matrix]] [[glycoprotein]] | |||
* secreted by [[Osteoclast|osteoclasts]] | |||
* may cause | |||
** ↑ [[Osteoclast]] activation | |||
** ↑ [[angiogenesis]] | |||
| | |||
* High levels may be associated with | |||
** extensive osteolytic [[Disease|diseas]] | |||
** advanced [[Multiple myeloma|disease]] ([[Multiple myeloma|myeloma]]) | |||
* High levels may also play a protective role against [[bone resorption]] in [[Multiple myeloma|myeloma]] patients with with [[MAF (gene)|maf]] [[gene]] [[translocations]]. | |||
|- | |||
|'''[[Interleukin 3]] [[IL-3|(IL-3]])''' | |||
* a bifunctional [[cytokine]] | |||
* stimulates [[osteoclast]] formation, primarily through induction of [[Activin|activin A]] production | |||
* inhibits [[osteoblast]] differentiation through participation of [[CD45]] + [[hematopoietic]] [[Cells (biology)|cells]]. | |||
| | |||
* Increased levels found in [[multiple myeloma]] | |||
* Thought to be secreted by [[T-lymphocytes]] through a complex interaction with [[Multiple myeloma|myeloma cells]] | |||
|- | |||
|'''[[Vascular endothelial growth factor]] ([[VEGF]])''' | |||
* important in [[angiogenesis]] and growth | |||
* may promote [[Osteoclasts|osteoclast]] [[differentiation]] by substituting for [[Colony stimulating factors|M-CSF]] | |||
| | |||
* Majority of [[Multiple myeloma|myeloma cells]] secrete [[VEGF]] | |||
* Increases survival and growth of [[Multiple myeloma|myeloma cells]] | |||
|- | |||
|'''[[Interleukin 6]] ([[IL-6]])''' | |||
* a multifunctional [[cytokine]] | |||
* involved in [[bone metabolism]] | |||
* stimulates [[osteoclast]] [[differentiation]] | |||
* stimulates the [[Phosphoinositide 3-kinase|PI3K]]/[[Akt]]/[[mTOR]] pathway. This pathway plays a role in regulation of expression of: | |||
** [[IL-6]] | |||
** [[Vascular endothelial growth factor|VEGF]] | |||
** [[osteopontin]] | |||
| | |||
* Increases survival of [[Multiple myeloma|myeloma cells]] | |||
* Stimulates [[Multiple myeloma|myeloma cells]] to produce and release [[vascular endothelial growth factor]] ([[VEGF]]) that further stimulates [[osteoclasts]] | |||
* correlates with [[bone turnover]] rate in [[multiple myeloma]] | |||
|- | |||
|'''[[Interleukin 17]] ([[Interleukin 17|IL-17]])''' | |||
* a pro-[[inflammatory]] [[cytokine]] | |||
* stimulates [[osteoclast]] activation | |||
| | |||
* Primarily secreted by [[T-helper cells]] ([[Th17]]). | |||
* Also secreted by [[T-lymphocytes]] and [[Natural killer cells|natural killer (NK) cells]] | |||
* Associated with osteolytic lesions in [[Multiple myeloma|myeloma]] models. | |||
|- | |||
|'''[[BAFF receptor|B cell-activating factor]] ([[BAFF receptor|BAFF]])''' | |||
'''Binding to its receptor → activation of [[NF-κB]] → ↑ [[Multiple myeloma|MM]] cell survival''' | |||
* a member of [[TNF|TNF superfamily]] | |||
* associated with increased [[osteoclast]] activation | |||
| | |||
* Secreted by [[bone marrow]] stromal cells (BMSCs), [[osteoclasts]], and [[Multiple myeloma|myeloma cells]] | |||
* Increased [[serum]] levels in [[multiple myeloma]] [[patients]] | |||
* Increases survival of [[Multiple myeloma|myeloma cells]] | |||
|- | |||
|'''[[Bruton's tyrosine kinase|Bruton’s tyrosine kinase]] ([[BTK]])''' | |||
'''[[Osteoclasts|Osteoclast]] precursors with CXC [[CXCR4|chemokine receptor type 4]] ([[CXCR4]]) and [[Bruton's tyrosine kinase|Bruton’s tyrosine kinase]] ([[Bruton's tyrosine kinase|BTK]]) expression → migration towards [[Stromal cell-derived factor-1|stromal cell-derived factor-1α]] ([[Stromal cell-derived factor-1|SDF-1α]]) → ↑ activation of [[BTK|Bruton’s tyrosine kinase]] ([[BTK]]) in [[Multiple myeloma|myeloma cells.]]''' | |||
* a nonreceptor [[tyrosine kinase]] | |||
* involved in [[B cell receptor]] signaling pathway | |||
* stimulates [[osteoclast]] [[differentiation]] | |||
| | |||
* Expressed by [[Multiple myeloma|myeloma cells]] | |||
* Correlates with [[CXCR4]] expression | |||
* Associated with progression, [[invasion]] through the [[extracellular matrix]] and the [[blood vessels]], and settling of [[Multiple myeloma|myeloma cells]] | |||
|- | |||
|'''[[Stromal cell-derived factor-1|Stromal cell-derived factor-1α]] ([[Stromal cell-derived factor-1|SDF-1α]])''' | |||
'''[[Osteoclasts|Osteoclast]] precursors with CXC [[CXCR4|chemokine receptor type 4]] ([[CXCR4]]) and [[Bruton's tyrosine kinase|Bruton’s tyrosine kinase]] ([[Bruton's tyrosine kinase|BTK]]) expression → migration towards [[Stromal cell-derived factor-1|stromal cell-derived factor-1α]] ([[Stromal cell-derived factor-1|SDF-1α]]) → ↑ activation of [[BTK|Bruton’s tyrosine kinase]] ([[BTK]]) in [[Multiple myeloma|myeloma cells.]]''' | |||
* a [[chemokine]] | |||
* stimulates [[osteoclasts]] by binding to [[CXCR4|CXC chemokine receptor type 4]] ([[CXCR4]]) | |||
| | |||
* Involved in migration of [[Multiple myeloma|myeloma cells]] | |||
* Plays a role in homing of [[Tumor cell|tumor cells]] | |||
|- | |||
|'''[[Annexin II]] ([[Annexin A2]])''' | |||
* a calcium-dependent phospholipid-binding member of the [[Annexin|annexin family]] | |||
* normally expressed by: | |||
** [[bone marrow]] stromal [[cells]] (BMSCs) | |||
** [[endothelial cells]] | |||
** [[Macrophages|mononuclear macrophages]] | |||
** [[osteoblasts]] | |||
** [[osteoclasts]] | |||
* Promotes | |||
** [[Mineralization of bone|mineralization]] | |||
** [[Osteoclasts|osteoclastogenesis]] | |||
| | |||
* Secreted by [[Multiple myeloma|myeloma cells]], [[bone marrow]] stromal [[cells]] (BMSCs), [[osteoblasts]], and [[osteoclasts]] | |||
* Up-regulated in [[multiple myeloma]] | |||
* Promotes | |||
** [[Multiple myeloma|myeloma cells]]' [[adhesion]] | |||
** growth of [[Multiple myeloma|myeloma cells]] | |||
** [[angiogenesis]] | |||
|- | |||
|'''PU.1''' | |||
* a [[Transcription|transcriptional]] activator | |||
* binds to the PU-box, a [[lymphoid]]-specific [[enhancer]] | |||
* may be associated with [[differentiation]] or activation of [[macrophages]] and [[B-cells]] | |||
* involved in [[osteoclasts]] formations | |||
* may modulate pre-[[RNA splicing|mRNA splicing]] | |||
| | |||
|} | |||
===== Decreased osteoblast activity ===== | |||
Inhibition of osteoblasts leading to decreased bone formation is now considered to be a critical event in bone disease in multiple myeloma. This inhibition leads to bone loss as well as inability to repair the osteoytic lesions caused by increased osteoclastic activity. Several pathways and factors have been associated with suppressed osteoblastic activity. Some of them are mentioned below in the table.<ref name="pmid29330358">{{cite journal |vauthors=Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA |title=Pathogenesis of bone disease in multiple myeloma: from bench to bedside |journal=Blood Cancer J |volume=8 |issue=1 |pages=7 |date=January 2018 |pmid=29330358 |pmc=5802524 |doi=10.1038/s41408-017-0037-4 |url=}}</ref><ref name="pmid18406675">{{cite journal |vauthors=Edwards CM, Zhuang J, Mundy GR |title=The pathogenesis of the bone disease of multiple myeloma |journal=Bone |volume=42 |issue=6 |pages=1007–13 |date=June 2008 |pmid=18406675 |pmc=2474770 |doi=10.1016/j.bone.2008.01.027 |url=}}</ref><ref name="pmid25187738">{{cite journal |vauthors=Hameed A, Brady JJ, Dowling P, Clynes M, O'Gorman P |title=Bone disease in multiple myeloma: pathophysiology and management |journal=Cancer Growth Metastasis |volume=7 |issue= |pages=33–42 |date=2014 |pmid=25187738 |pmc=4133035 |doi=10.4137/CGM.S16817 |url=}}</ref> | |||
{| class="wikitable" | |||
|+ | |||
!Molecular pathways and factors associated with osteoblastic activity | |||
!Association with multiple myeloma | |||
|- | |||
|'''[[Wnt signaling pathway|Wingless and integration-1]] ([[Wnt signaling pathway|Wnt]]) signaling''' | |||
* '''[[Canonical sequence|Canonical]] pathway''' | |||
'''Activation of [[Wnt signaling pathway|Wnt pathway]] → binding of [[Wnt signaling pathway|Wnt ligands]] to [[Wnt signaling pathway|Wnt co-receptors]] [[LRP5]]/[[LRP6|6]] and one trans-membrane receptor of the FDZ family →''' | |||
'''formation of [[DVL1|DVL]]–Axin–[[FRAT1]]–[[GSK-3β]] complex → [[Translocations|translocation]] of [[β-catenin]] from [[cytoplasm]] to [[nucleus]] →''' | |||
'''activation of [[T cell]] factor/lymphoid enhancer factor (TCF/LEF) transcription factors → ↑ [[bone]] formation and ↓ [[bone resorption]]''' | |||
* '''Non-canonical [[Wnt signaling pathway|WNT]]–planar cell polarity (WNT–[[PCP (complexity)|PCP]]) pathway''' | |||
'''Formation of [[Wnt signaling pathway|WNT]] [[ligand]]–[[ROR2|receptor tyrosine kinase-like orphan receptor 2]] ([[ROR2]]) or the receptor-like tyrosine kinase (RYK)–FZD–DVL complex''' | |||
'''→ activation of one of these 3 pathways: 1) Disheveled-associated activator of morphogenesis 1 (DAAM1)–RHO–RHO-associated kinase (ROCK) pathway; 2) RAC–Jun kinase (JNK)–RUNX2 pathway; [[Wnt signaling pathway|WNT]]–Ca<sup>2+</sup> pathway''' | |||
* Binding of [[PTH]] to [[PTH receptor 1|PTH1 receptor]] may result in activation of [[Wnt signaling pathway|Wnt pathway]] | |||
* This pathway is also involved in [[Cell cycle|cell-cycle]] promotion and cell growth | |||
* | |||
|'''Dickkopf 1 (Dkk-1)''' | |||
* an [[extracellular]] [[inhibitor]] of this pathway expressed by [[Osteoblast|osteoblasts]] and [[bone marrow]] stromal cells, is thought to play the crucial role in [[Osteoblast|osteoblastic]] inhibition in [[multiple myeloma]]. | |||
* The [[serum]] levels and the expression of Dickkopf 1 (Dkk-1) by [[bone marrow]] cells were found to increased in [[patients]] with [[Multiple myeloma|multiple myeloma.]] | |||
'''[[Sclerostin]]''' | |||
* secreted by [[osteocytes]], [[sclerostin]] is a [[cysteine]] knot-containing [[protein]] | |||
* activates the [[Caspase|caspase pathway]], resulting in [[apoptosis]] of mature [[osteoblasts]] | |||
* binds to [[LRP5]]/[[LRP6|6]] transmembrane [[receptors]] preventing the [[Wnt signaling pathway|Wnt]] ligand- [[LRP5]]/[[LRP6|6]] transmembrane [[receptors]] binding, that leads to inhibition of [[Wnt signaling pathway]] | |||
* promotes osteoclastogenesis by increasing [[RANKL]]/[[OPGs|OPG]] ratio | |||
* elevated levels have been found in all phases of [[multiple myeloma]] and is considered to be a worse [[Prognostic|prognostic factor]]. Furthermore, [[Multiple myeloma|myeloma cells]] have been shown to secrete [[sclerostin]] | |||
'''[[SFRP2|Secreted frizzled related protein-2]] ([[SFRP2|sFRP-2]])''' | |||
* prevents binding of [[Wnt signaling pathway|Wnt ligands]] to [[frizzled]], a [[membrane]] bound [[receptor]] | |||
* some studies suggest that [[Multiple myeloma|myeloma cells]] express [[SFRP2|sFRP-2]] that [[Antagonists|antagonizes]] [[bone]] formation | |||
'''[[Periostin]]''' | |||
* produced by [[bone marrow]] stromal [[cells]], [[periostin]] is an [[Adhesion molecule|adhesion protein]] of fasciclin family | |||
* thought to activate the [[integrin]]–[[AKT]]–FAK–[[β-catenin]] pathway | |||
* although the role is still unclear, elevated levels have been found in almost all the patients presenting with [[multiple myeloma]] | |||
'''[[Runx2|Runt-related transcription factor 2]] ([[Runx2]])/corebinding factor runt domain alpha subunit 1 (CBFA1)''' | |||
* a [[transcription factor]] that is a part of the non-canonical [[Wnt signaling pathway|WNT-signaling pathway]] | |||
* [[Multiple myeloma|myeloma cells]] decrease [[osteoblast]] [[differentiation]] by inhibiting [[RUNX2]] activity in BMSCs and [[osteoblast]] [[precursor]] [[cells]] | |||
* | |||
* | |||
|- | |||
|'''[[Ephrin B1|EphrinB2]]/[[Eph receptor|EphB4]] signaling pathway''' | |||
'''Binding of ligands called [[Ephrin|ephrins]] (Eph receptor-interacting proteins) → activation of two cascades: 1) the forward signaling → ↑ [[osteoblast]]''' | |||
'''[[differentiation]] by downregulating [[RhoA]]; 2) the reverse signaling → ↓ [[osteoclast]] [[differentiation]] by ↓ Fos and Nfatc1 [[transcription]]''' | |||
* [[Ephrin|EphrinB2]] is found in [[osteoclasts]] | |||
* [[Eph receptor|EphB4]] is found in [[Osteoblast|osteoblasts]] and [[bone marrow]] stromal [[cells]] | |||
|[[Multiple myeloma|Multiple myeloma patients]] have decreased expression of both [[Ephrin B2|EphrinB2]] and [[Eph receptor|EphB4]] in [[bone marrow]] stromal [[cells]]. | |||
|- | |||
|'''[[Transforming growth factor-β]] ([[TGF-β]])''' | |||
* a ubiquitous, multifunctional [[growth factor]] | |||
* produced by [[osteocytes]] and [[Osteoblast|osteoblasts]] and is activated by [[osteoclasts]] | |||
* primary effect in [[bone marrow]] is the inhibition of terminal [[differentiation]] of [[osteoblasts]] | |||
|Inhibition of [[Transforming growth factor-β|Transforming Growth Factor-β]] have been shown to prevent [[Multiple myeloma|myeloma cells]] to block [[Osteoblast|osteoblastic]] [[differentiation]]. | |||
|- | |||
|'''[[Bone morphogenetic proteins]] ([[Bone morphogenetic protein|BMPs]])''' | |||
* belong to [[TGFβ]] superfamily | |||
* promote osteoblastogenesis through [[Smad]]-dependent and [[Smad]]-independent pathways | |||
|[[Multiple myeloma|Myeloma cells]] express negative regulators of [[bone morphogenetic proteins]] that result in decreased activity of these [[proteins]] which in turn causes decreased osteoblastogenesis. | |||
|- | |||
|'''[[Hepatocyte growth factor]] ([[Hepatocyte growth factor|HGF]])''' | |||
* inhibits [[BMP]]-induced osteoblastogenesis, thought to be mediated by inhibition of [[Cell nucleus|nuclear]] [[Translocations|translocation]] of receptor-activated [[SMAD (protein)|SMADs]] | |||
| | |||
* Produced by [[Multiple myeloma|myeloma cells]] | |||
* Increased levels have been shown to correlate with worse [[prognosis]] | |||
|- | |||
|'''[[Interleukin 3]] ([[IL-3]])''' | |||
* inhibits [[BMP]]-induced osteoblastogenesis, thought to be mediated indirectly by increasing [[CD45]]+ [[hematopoietic]] [[cells]] | |||
|Elevated levels had been demonstrated in [[bone marrow]] of [[Multiple myeloma|myeloma patients]]. | |||
|- | |||
|'''[[Interleukin 7]] ([[IL-7|IL-7)]]''' | |||
* Causes | |||
** increased osteoclastic activity | |||
** decreased [[Osteoblast|osteoblasts]] stimulation and maturation | |||
** decreased [[Runx2]]/CBFA1 activity | |||
| | |||
|- | |||
|'''[[GFI1|Growth factor independence-1]] ([[GFI1]])''' | |||
* a [[Repressor|transcriptional repressor]] that causes decreased expression of [[RUNX2]] | |||
|[[Bone marrow]] stromal [[cells]] (BMSCs) in [[Multiple myeloma|myeloma patients]] have been shown to over-express [[GFI1|growth factor independence-1]] ([[GFI1]]). | |||
|- | |||
|'''[[Tumor necrosis factor-alpha|Tissue necrosis factor-α]] ([[TNF-α]])''' | |||
* inhibits osteoblast differentiation through | |||
** inhibits osteoblast precursor recruitment | |||
** suppresses RUNX2 and its transcriptional co-activator, TAZ | |||
|[[Tumor necrosis factor-alpha|Tissue necrosis factor-α]] ([[TNF-α]]) levels are increased in [[multiple myeloma]] and it is thought to play a complex role in [[pathogenesis]] of [[Multiple myeloma|multiple myeloma.]] | |||
|- | |||
|'''[[Adiponectin]]''' | |||
[[adipocyte]]-derived [[hormone]] thought to act on [[Osteoblast|osteoblasts]] and [[osteoclasts]] | |||
|Increased [[adiponectin]] secretion through [[pharmacologic]] interventions have shown to decrease osteolytic lesions in [[Multiple myeloma|myeloma]]. | |||
|} | |||
===== Renal involvement in multiple myeloma ===== | |||
Renal involvement is common in multiple myeloma as up to 50% of the patients go on to develop kidney disease at some point during the disease course. Half of these patients recover kidney function while the rest of them develop chronic kidney dysfunction. The causes of renal involvement in multiple myeloma are multiple and diverse. They can broadly be classified into Ig dependent and Ig independent mechanisms. Some of them have been discussed below in the table and then described briefly.<ref name="pmid18528426">{{cite journal |vauthors=Dimopoulos MA, Kastritis E, Rosinol L, Bladé J, Ludwig H |title=Pathogenesis and treatment of renal failure in multiple myeloma |journal=Leukemia |volume=22 |issue=8 |pages=1485–93 |date=August 2008 |pmid=18528426 |doi=10.1038/leu.2008.131 |url=}}</ref><ref name="pmid24877060">{{cite journal |vauthors=Mussap M, Merlini G |title=Pathogenesis of renal failure in multiple myeloma: any role of contrast media? |journal=Biomed Res Int |volume=2014 |issue= |pages=167125 |date=2014 |pmid=24877060 |pmc=4022292 |doi=10.1155/2014/167125 |url=}}</ref><ref name="pmid23868898">{{cite journal |vauthors=Heher EC, Rennke HG, Laubach JP, Richardson PG |title=Kidney disease and multiple myeloma |journal=Clin J Am Soc Nephrol |volume=8 |issue=11 |pages=2007–17 |date=November 2013 |pmid=23868898 |pmc=3817918 |doi=10.2215/CJN.12231212 |url=}}</ref> | |||
{| class="wikitable" | |||
|+ | |||
! colspan="2" |Ig-dependent renal injury | |||
! colspan="2" |Ig-independent renal injury | |||
|- | |||
!Cause | |||
!Notes | |||
!Cause | |||
!Notes | |||
|- | |||
| '''Cast [[nephropathy]] ([[Multiple myeloma|myeloma]] [[kidney]])''' | |||
|Risk factors | |||
* [[light chain]] [[Multiple myeloma|myeloma]] with >10 g/d [[monoclonal]] [[Immunoglobulin|Ig]] excretion | |||
* volume depletion | |||
* [[sepsis]] | |||
* [[medications]] | |||
* [[Immunoglobulin|Ig]] deposition is primarily in the [[renal tubules]] | |||
| '''[[Hypovolemia|Volume depletion]]''' | |||
|May cause | |||
* pre-renal [[azotemia]] | |||
* [[acute tubular necrosis]] and/or contribute to cast nephropathy | |||
|- | |||
| '''[[Monoclonal]] [[Immunoglobulin|Ig]] deposition [[disease]]''' | |||
| | |||
* May be associated with systemic [[syndrome]] | |||
* [[Immunoglobulin|Ig]] deposition may be demonstrated in either [[Renal tubules|tubules]] or [[glomeruli]] but typically not in both | |||
| '''[[Sepsis]]''' | |||
|May cause | |||
* pre-renal [[azotemia]] | |||
* [[acute tubular necrosis]] and/or contribute to cast nephropathy | |||
|- | |||
| '''[[Amyloidosis AL|Light chain amyloidosis]] ([[Amyloidosis AL|AL]])''' | |||
| | |||
* May be associated with systemic [[syndrome]] | |||
* often with [[Nephrotic syndrome|nephrotic]]-range [[albuminuria]] and [[Light chain|λ-light chains]] | |||
* [[amyloid]] deposition is primarily in the [[glomeruli]] | |||
| '''[[Hypercalcemia]]''' | |||
|May result in [[acute kidney injury]] directly or contribute to cast nephropathy | |||
|- | |||
| '''[[Glomerulonephritis]]''' | |||
|Following types have been demonstrated | |||
* [[Membranoproliferative GN|membranoproliferative]] | |||
* [[Glomerulonephritis|diffuse proliferative]] | |||
* [[Crescentic glomerulonephritis|crescentic]] | |||
* cryoglobulinemic | |||
| '''[[Tumor lysis syndrome]]''' | |||
|Caused by [[uric acid]] or [[phosphate]] nephropathy | |||
|- | |||
| [[Tubulointerstitial nephritis|'''Tubulointerstitial''' '''nephritis''']] | |||
|May be caused be either [[Immunoglobulin|Ig]]-dependent or [[Immunoglobulin|Ig]]-independent mechanisms | |||
| '''Direct parenchymal invasion by [[plasma cells]]''' | |||
|Rare cause. Association with advanced or aggressive [[multiple myeloma]] | |||
|- | |||
| '''[[Minimal change disease]]''' | |||
|Often with [[albuminuria]] and [[light chain]] [[proteinuria]] | |||
| '''[[Pyelonephritis]]''' | |||
|Rare cause. [[Pathogenesis]] is typically multifactorial and may include: | |||
* [[immunodeficiency]] | |||
* deficient [[Immunoglobulin|Ig]] | |||
* [[chemotherapy]] | |||
|- | |||
| '''[[Hyperviscosity syndrome]]''' | |||
|Seen primarily in cases of [[IgA]], [[IgG|IgG3]], or [[IgM]] [[Multiple myeloma|myeloma]] | |||
| rowspan="5" | '''Medication toxicity''' | |||
| rowspan="5" |Following [[drugs]] may cause kidney injury: | |||
* [[Zoledronate]]: May cause acute [[Kidney|renal]] failure in rare cases | |||
* [[Pamidronate]]: May be associated with collapsing focal and segmental [[glomerulosclerosis]] in rare cases | |||
* [[Nonsteriodal anti-inflammatory drugs]] | |||
* [[Angiotensin converting enzyme inhibitors|Angiotensin converting enzyme inhibitor]] | |||
* [[Angiotensin receptor blocker]] | |||
* [[Loop diuretics]] | |||
* [[Iodinated contrast]] | |||
|- | |||
| '''[[Henoch-Schönlein purpura|Henoch–Scholein purpura]]/[[IgA nephropathy]]''' | |||
|Associated with [[IgA]] [[Multiple myeloma|myeloma]] | |||
|- | |||
| '''Immunotactoid glomerulopathy (and possibly fibrillary GN)''' | |||
|Rare conditions. The association between fibrillary disease and [[Paraprotein|paraproteins]] is is not understood at this point. | |||
|- | |||
| '''[[Thrombotic microangiopathy]] ([[TMA]])''' | |||
|[[Endothelium|Endothelial]] injury caused by [[paraprotein]] leads to [[Thrombotic microangiopathy|thrombotic microangiopathy.]] | |||
|- | |||
| '''[[Membranous glomerulonephritis|Membranous glomerulopathy]]''' | |||
| | |||
|} | |||
====== '''Cast nephropathy (myeloma kidney)''' ====== | |||
* Normally circulating [[monoclonal]] [[Light chain|free light chains]] are relatively freely filtered through the [[glomerulus]] and a [[receptor]]-mediated process leads to their [[endocytosis]] in [[Proximal tubule cell|proximal tubule cells]]. They bind to the tandem [[scavenger receptor]] system [[cubilin]]/megalin, followed by [[endocytosis]] through the [[clathrin]]-dependent [[Endosome|endosomal]]/[[Lysosome|lysosomal]] pathway. They are then degraded within [[lysosomes]]. | |||
* In [[multiple myeloma]], [[Multiple myeloma|myeloma cells]] produce excess of [[monoclonal]] [[Light chain|free light chains]] that overwhelms the capacity of the [[Proximal tubule cell|proximal tubule cells]] to absorb and [[Catabolism|catabolize]] them. This leads to large amounts of [[monoclonal]] [[Light chain|free light chains]] reaching [[Distal convoluted tubule|distal renal tubules]]. | |||
* In distal renal tubules, they interact with Tamm-Horsfall protein (uromodulin), a glycoprotein produced in the medullary thick ascending limb of the loop of Henle in kidneys. This interaction leads to formation of myeloma casts. | |||
* These casts block the [[glomerular]] flow, compromising [[renal]] function and may lead to [[Proximal tubule|proximal tubular]] [[atrophy]]. This may also contribute to interstitial fibrosis. | |||
* This blockage leads to increased luminal pressure, resulting in decreased [[blood flow]] and causing further [[renal]] injury. | |||
* The excessive amounts of [[monoclonal]] [[Light chain|free light chains]] being absorbed in proximal renal tubules leads to induction of apoptosis and DNA damage in proximal renal tubules. | |||
* Free light chains also leads to increased pro-[[inflammatory]] [[cytokines]] such as [[Interleukin 6|IL-6]], [[MCP-1|macrophage chemoattractant protein-1]] ([[MCP-1]]), and [[Tumor necrosis factor-alpha|tumor necrosis factor ''α'']] ([[TNFα|TNF''α'']]) through [[transcription factors]] [[NF-kB|nuclear factor kappa B]] ([[NFκB|NF''κ''B]]) and [[Mitogen-activated protein kinase|mitogen-activated protein kinases]]. | |||
* All these events leads to kidney injury and inability of [[renal]] [[cells]] to repair the injury. On histopathological studies, [[Tubulointerstitial diseases of the kidney|chronic tubulointerstitial nephropathy]] with marked [[tubular]] [[atrophy]], laminated casts in the [[Renal tubule|tubules]], and interstitial fibrosis may be observed. | |||
{| class="wikitable" | |||
|+ | |||
!Factors associated with increased risk | |||
of AKI or CKD in multiple myeloma | |||
!Mechanism of injury | |||
|- | |||
|[[Hypovolemia|Volume depletion]] | |||
|Increased [[Light chain|FLC]] concentration | |||
Decreased [[Glomerular filtration rate|GFR]] | |||
Pre-renal [[azotemia]] | |||
|- | |||
|[[Hypercalcemia]] | |||
|[[Renal]] [[vasoconstriction]] | |||
Decreased [[GFR]] | |||
Pre-[[renal]] [[azotemia]] | |||
|- | |||
|[[Iodinated contrast|Iodinated contrast media]] | |||
|[[Nephrotoxicity]] | |||
|- | |||
|[[Nonsteroidal anti-inflammatory drugs]] | |||
|[[Renal]] [[vasoconstriction]] | |||
Decreased [[GFR]] | |||
[[Renal medulla|Medullary]] [[hypoxia]] | |||
|- | |||
|[[Diuretics]] | |||
|Increased [[sodium chloride]] concentration | |||
Increased cast formation | |||
|- | |||
|[[Aminoglycosides]] | |||
|[[Renal]] [[vasoconstriction]] | |||
Decreased GFR | |||
|- | |||
| colspan="2" |[[Hyperuricemia]] | |||
|- | |||
| colspan="2" |[[Comorbidities]] such as [[chronic kidney disease]], [[diabetes]], [[aging]], [[hypertension]], and [[cardiovascular disease]]. | |||
|} | |||
====== Hypercalcemia ====== | |||
* [[Hypercalcemia]] in [[multiple myeloma]] primarily results from increased [[Osteoclasts|osteoclastic]] [[bone resorption]]. It is the second most common cause of [[renal]] injury in [[Multiple myeloma|myeloma patients]]. | |||
* [[Hypercalcemia]] leads to intra-[[Nephron|tubular]] [[calcium]] deposition and [[vasoconstriction]] in [[Renal blood flow|renal vasculature]]. The resultant decrease in [[GFR]] not only causes [[renal]] injury by itself but also favors cast formation. | |||
* By impairing [[renal]] concentrating ability through resistance to [[anti-diuretic hormone]], it may also cause [[nephrogenic diabetes insipidus]]. [[Polyuria]] may develop, leading to [[hypovolemia]]. Net effect is decreased [[GFR]], concentrated [[urine]], reduced [[urine]] flow, and [[Azotemia|pre-renal azotemia]]. | |||
====== Light-chain glomerulopathy ====== | |||
* Deposition of [[immunoglobulins]] either in the form of [[amyloid]] or non-[[amyloid]], typically resulting in non-selective [[proteinuria]]. | |||
* [[Amyloid]] deposits are fibrillar structures, consist of variable region fragments of the [[light chain]], and are seen primarily within the [[glomeruli]]. | |||
* Some [[patients]] may have predominant [[vascular]] [[amyloid]] deposits rather than [[Glomerulus|glomerular]] deposits and present with [[renal failure]] instead of [[nephrotic syndrome]]. | |||
====== Light chain deposition disease (LCDD) ====== | |||
* Non-fibrillar, granular deposits are topically observed in [[Mesangial cells|mesangial]] area along with thickening of [[basement membrane]] which resembles [[Membranoproliferative glomerulonephritis|type II membranoproliferative glomerulonephritis]] or [[diabetic glomerulopathy]]. The [[light chain]] deposits may also be observed in [[renal]] [[vasculature]] in addition to [[Mesangial cell|mesangial]] area. | |||
* Typical clinical picture is that of [[nephrotic syndrome]] but [[renal]] impairment is more severe with almost all patients developing [[renal failure]]. | |||
===== Immune dysfunction in multiple myeloma ===== | |||
* [[Immunodeficiency]] is an important characteristic of [[multiple myeloma]] and is thought to associated with increased risk for [[infections]], and affects [[disease]] progression and response to treatment.<ref name="pmid23176722">{{cite journal |vauthors=Borrello I |title=Can we change the disease biology of multiple myeloma? |journal=Leuk. Res. |volume=36 Suppl 1 |issue= |pages=S3–12 |date=November 2012 |pmid=23176722 |pmc=3698609 |doi=10.1016/S0145-2126(12)70003-6 |url=}}</ref><ref name="pmid">{{cite journal |vauthors=Cook G, Campbell JD |title=Immune regulation in multiple myeloma: the host-tumour conflict |journal=Blood Rev. |volume=13 |issue=3 |pages=151–62 |date=September 1999 |pmid= |doi=10.1054/blre.1999.0111 |url=}}</ref> | |||
* The typical clinical picture is that of global [[immunosuppression]]. The interaction between [[Multiple myeloma|myeloma]] cells and [[bone marrow]] stromal [[cells]] and [[Cytokine|cytokines]] produced as a result of this interaction are thought to play a key role.<ref name="pmid23176722">{{cite journal |vauthors=Borrello I |title=Can we change the disease biology of multiple myeloma? |journal=Leuk. Res. |volume=36 Suppl 1 |issue= |pages=S3–12 |date=November 2012 |pmid=23176722 |pmc=3698609 |doi=10.1016/S0145-2126(12)70003-6 |url=}}</ref><ref name="pmid">{{cite journal |vauthors=Cook G, Campbell JD |title=Immune regulation in multiple myeloma: the host-tumour conflict |journal=Blood Rev. |volume=13 |issue=3 |pages=151–62 |date=September 1999 |pmid= |doi=10.1054/blre.1999.0111 |url=}}</ref><ref name="pmid16966269">{{cite journal |vauthors=Schütt P, Brandhorst D, Stellberg W, Poser M, Ebeling P, Müller S, Buttkereit U, Opalka B, Lindemann M, Grosse-Wilde H, Seeber S, Moritz T, Nowrousian MR |title=Immune parameters in multiple myeloma patients: influence of treatment and correlation with opportunistic infections |journal=Leuk. Lymphoma |volume=47 |issue=8 |pages=1570–82 |date=August 2006 |pmid=16966269 |doi=10.1080/10428190500472503 |url=}}</ref><ref name="pmid8634441">{{cite journal |vauthors=Urashima M, Ogata A, Chauhan D, Hatziyanni M, Vidriales MB, Dedera DA, Schlossman RL, Anderson KC |title=Transforming growth factor-beta1: differential effects on multiple myeloma versus normal B cells |journal=Blood |volume=87 |issue=5 |pages=1928–38 |date=March 1996 |pmid=8634441 |doi= |url=}}</ref> | |||
* Following abnormalities have been demonstrated in myeloma patients:<ref name="pmid16966269">{{cite journal |vauthors=Schütt P, Brandhorst D, Stellberg W, Poser M, Ebeling P, Müller S, Buttkereit U, Opalka B, Lindemann M, Grosse-Wilde H, Seeber S, Moritz T, Nowrousian MR |title=Immune parameters in multiple myeloma patients: influence of treatment and correlation with opportunistic infections |journal=Leuk. Lymphoma |volume=47 |issue=8 |pages=1570–82 |date=August 2006 |pmid=16966269 |doi=10.1080/10428190500472503 |url=}}</ref><ref name="pmid14717778">{{cite journal |vauthors=Mozaffari F, Hansson L, Kiaii S, Ju X, Rossmann ED, Rabbani H, Mellstedt H, Osterborg A |title=Signalling molecules and cytokine production in T cells of multiple myeloma-increased abnormalities with advancing stage |journal=Br. J. Haematol. |volume=124 |issue=3 |pages=315–24 |date=February 2004 |pmid=14717778 |doi= |url=}}</ref><ref name="pmid8634441">{{cite journal |vauthors=Urashima M, Ogata A, Chauhan D, Hatziyanni M, Vidriales MB, Dedera DA, Schlossman RL, Anderson KC |title=Transforming growth factor-beta1: differential effects on multiple myeloma versus normal B cells |journal=Blood |volume=87 |issue=5 |pages=1928–38 |date=March 1996 |pmid=8634441 |doi= |url=}}</ref> | |||
** Decreased numbers of [[Natural killer cell|natural killer cells]], [[B-cells]], and [[memory T cells]] | |||
** Decreased non-[[Multiple myeloma|myeloma]] [[immunoglobulins]] | |||
** Decreased expression of cell-surface markers on [[CD4+]] and [[CD8+ T cells|CD8+]] [[T cells]] such as [[CD28]] and CD152. These markers are associated with [[Cell signaling|cell-signaling]]. | |||
** Abnormal [[signal transduction]] in [[T-cells]] | |||
** Abnormalities in [[Cytokines|cytokine]] production | |||
** Abnormalities in [[antibodies]] response and production | |||
** Aberrations in [[B-cells]] [[maturation]] | |||
** Defects in [[Natural killer cell|natural killer cells]] and [[antigen presenting cells]] | |||
==Gross Pathology== | |||
<gallery> | |||
Image:Vertebras in multiple myeloma 0001.jpg| Vertebrae in multiple myeloma <br> (Image courtesy of Melih Aktan M.D.) | |||
Image:Calvarium in multiple myeloma.jpg | Calvarium in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | |||
</gallery> | |||
==Microscopic Pathology== | |||
On microscopic histopathological analysis, multiple myeloma is characterized by the following:<ref name="patho">Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015</ref> | |||
:*Abundant eosinophilic [[cytoplasm]] | |||
:*Eccentrically placed [[nucleus]] | |||
:*Clock face morphology of the nucleus due to [[chromatin]] clumps around the edges | |||
:*Russell bodies which are eosinophilic, large (10-15 micrometres), homogenous immunoglobulin-containing inclusions | |||
:*Dutcher bodies which are [[PAS stain]] +ve intranuclear crystalline rods | |||
*Shown below is a series of microscopic images seen in multiple myeloma: | |||
<gallery> | <gallery> | ||
Image:Bone marrow aspiration in multiple myeloma 0001.jpg|Bone marrow aspiration in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | Image:Bone marrow aspiration in multiple myeloma 0001.jpg|Bone marrow aspiration in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | ||
Image:Bone marrow biopsy in multiple myeloma 0001.jpg|Bone marrow biopsy in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | Image:Bone marrow biopsy in multiple myeloma 0001.jpg|Bone marrow biopsy in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | ||
Image:Bone marrow in multiple myeloma 0001.jpg|Bone marrow in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | Image:Bone marrow in multiple myeloma 0001.jpg|Bone marrow in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | ||
Image:Bone marrow in multiple myeloma 0002.jpg|Bone marrow in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | Image:Bone marrow in multiple myeloma 0002.jpg|Bone marrow in multiple myeloma. <br> (Image courtesy of Melih Aktan M.D.) | ||
File:Multiple myeloma intermed mag.jpg|Multiple myeloma slide with intermediate magnification<ref name="patho">Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015</ref> | |||
File:Multiple myeloma.jpg|Multiple myeloma slide with high magnification<ref name="patho">Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015</ref> | |||
Russell bodies.jpg|Multiple myeloma slide with russell bodies<ref name="patho">Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015</ref> | |||
</gallery> | </gallery> | ||
==References== | ==References== | ||
{{Reflist|2}} | {{Reflist|2}} | ||
[[Category: | [[Category:Medicine]] | ||
[[Category:Hematology]] | [[Category:Hematology]] | ||
[[Category: | [[Category:Neurology]] | ||
[[Category:Neurosurgery]] | |||
[[Category:Oncology]] | [[Category:Oncology]] | ||
[[Category:Up-To-Date]] | |||
[[Category:Surgery]] | |||
Latest revision as of 22:47, 29 July 2020
|
Multiple myeloma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Multiple myeloma pathophysiology On the Web |
|
American Roentgen Ray Society Images of Multiple myeloma pathophysiology |
|
Risk calculators and risk factors for Multiple myeloma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1] Associate Editor(s)-in-Chief: Hannan Javed, M.D.[2]; Haytham Allaham, M.D. [3]; Shyam Patel [4]
Overview
Multiple myeloma, a disorder of clonal late B-cells, arises from post-germinal center plasma cells that are normally involved in production of human immunoglobulins.[1][2][3] Although the exact pathogenesis and the stage at which myeloma cells arise from post-germinal B-cells remain unclear, a variety of factors have been implicated in pathogenesis of multiple myeloma. Of these, chromosomal abnormalities are thought to be the most important. It has been suggested that all cases of multiple myeloma pass through MGUS. Renal involvement by multiple myeloma is catergorized into three entities: light chain cast nephropathy, monoclonal immunoglobulin deposition disease, and amyloidosis. Osseous involvement by multiple myeloma is based on cytokine and cellular interactions that lead to bone breakdown. On microscopic histopathological analysis, abundant eosinophilic cytoplasm, eccentrically placed nucleus, and Russell bodies are characteristic findings of multiple myeloma.[4]
Pathophysiology
Normal physiology and development of plasma cells
- Plasma cells are terminally differentiated B-cells which function to produce immunoglobulins. plasma cells arise from B lymphocytes through a series of events. These events take place in bone marrow and secondary lymphoid tissues.
- These events include immunoglobulin heavy-chain (IgH) VDJ gene rearrangement, migration into lymphoid tissues, somatic hypermutation (SMH) in the IgH and light-chain genes, antigen selection, switch in Ig class from IgM to IgG, IgA, IgD, or IgE, migration back to bone marrow and terminal differentiation into plasma cells.[5] [6][7]
Stem cells → Pre-B cells → Immature B-cells → Mature B-cells (naïve) → Activated B-cells → Memory B-cells and Plasmablasts → Plasma cells
- During these events, B cells express variety of surface markers which may be used to denote their developmental stage such as CD19, CD20, CD27, CD38, CD10, CD138. These markers and IgH chain gene sequences are important to define the nature of myeloma cells.[5][6][7]
- Plasma cells secrete antibodies which function in humoral immunity.[5][6] [7]
- Plasma cells are typically polyclonal and can respond to a variety of antigens, which helps combat infections. Under normal circumstances, there is no monoclonality amongst the plasma cell population in a person. These plasma cells are functionally intact in their ability to contribute to humoral immunity.[5][6][7]
- Transcription factors such as interferon regulatory factor 4 (IRF4), BCL6, B-lymphocyte-induced maturation protein 1 (BLIMP1, also known as PRDM1), paired box gene 5 (PAX5) and X box binding protein 1 (XBP1) play an important role in differentiation and survival of plasma cells.[8][9][10]
Interferon regulatory factor 4 (IRF4) → Down-regulation of BCL6 → Up-regulation of B-lymphocyte-induced maturation protein 1 (BLIMP1) → Down-regulation of paired box gene 5 (PAX5) and Up-regulation of X box binding protein 1 (XBP1).[11][12][13]
For more information on plasma cells, click here.
Normal physiology and development of Immunoglobulins
- Immunoglobulins (also known as Antibodies) are proteins that are found in blood or other bodily fluids of vertebrates, and are used by the immune system to identify and neutralize foreign objects, such as bacteria and viruses.
- They are made of a few basic structural units called chains; each antibody has two large heavy chains H and two small light chains L. There are several different types of antibody heavy chains, and several different kinds of antibodies, which are grouped into different isotypes based on which heavy chain they possess.[14][15]
- Five different antibody isotypes are known in mammals, which perform different roles, and help direct the appropriate immune response for each different type of foreign object they encounter.[14][15]
- Class switch recombination (CSR), a region specific deletion recombination process, generates different immunoglobulin (Ig) isotypes by replacing one switch region with another. This process leads to enhanced functionality of immunoglobulins.[16][17]
- Class switch recombination (CSR), just like somatic hypermutation (SMH), requires the expression of activation-induced deaminase (AID) and both are dependent on creation of double-stranded DNA breaks (DSB) in the Ig loci.[18][19][20]
For more information on immunoglobulins, click here.
Pathogenesis
The pathogenesis of multiple myeloma is complex and probably is a result of multiple and multi-step oncogenic events such as hyperdiploidy and deregulation of cyclin D1, and interaction of myeloma cells with marrow environment. Recently it has been suggested that all cases of multiple myeloma pass through an MGUS phase. The events surrounding the progression of MGUS into multiple myeloma are not well-defined but environmental and genetic factors have been proposed to have an association. A brief description of events thought to play a role in pathogenesis of multiple myeloma is given here.
Biology of myeloma cells
- Myeloma cells are malignant plasma cells which exhibit the morphology of mature plasma cells or plasmablasts. Majority of these cells seem to be mature, differentiated and quiescent, appearing to be without long-term proliferative potential.[21][22]
- Cells with similar morphology to mature B-cells but with immunoglobulin gene sequences and idiotype similar to myeloma cells have also been found in myeloma patients, both in bone marrow and the peripheral blood.[23][22]
- Current hypothesis is the presence of functional heterogeneity in myeloma cells with only a minor group of specialized myeloma cells exhibiting clonogenic growth potential. Although studies have shown the presence of myeloma cells sub-populations with distinct phenotypes and functionality in multiple myeloma such as presence of CD138+ and CD138− sub-populations, the phenotype of these so-called clonogenic cells is yet to be determined. [23][22][24]
- Myeloma cells express surface markers associated with plasma cells such as CD138, natural killer (NK) cells such as CD56/NCAM, T cells such as CD28, and sometimes the pan-B-cell marker CD20. Presence of CD19 and CD20 on sub-population of myeloma cells may suggest either early-lineage precursor for myeloma cells or possible de-differentiation of myeloma cells.[5][22][25][26][27]
- Myeloma cells show complex chromosomal abnormalities and mutations. Studies have demonstrated the presence of translocations in up to 75% of the myeloma cases.[5]
Environmental and hereditary factors
- The Key environmental and hereditary factors thought to confer a greater risk of developing multiple myeloma or play a part in pathogenesis are summarized in the table below.[8][28][29][30][31][32][33][34][35][36][37]
| Environmental and hereditary risk factors |
|---|
|
| * Likely influenced by environmental and behavioral confounding factors. |
Chromosomal aberrations
- Two major sets of chromosomal abnormalities seen in multiple myeloma are translocations with extensive IgH rearrangements and numerical aberrations, either trisomy or monosomy.[8][5]
Translocations
- Majority of translocations in multiple myeloma take place through class switch recombination (CSR). But few may occur through DH–JH recombination, possibly in early stages of B-cell development such as pre-B cell stage.[8][38]
- Majority of these translocations put oncogenes such as cyclin D1 (CCND1), CCND3, fibroblast growth factor receptor 3 (FGFR3), multiple myeloma SET domain (MMSET; also known as WHSC1), MAF and MAFB under the influence of enhancers present at Ig loci. .[5][8][39][40]
- These translocations cause over-expression of these oncogenes that in turn leads to dysregulation of cell-cycle such as increased cell survival, growth, progression and DNA repair. One of the key abnormality being the increased G1/S transition during the cell-cycle.[8][41]
- Many other translocations may occur in later phases of multiple myeloma. One important example is translocation of MYC, an oncogene that encodes transcription factors.[8][42][43]
Hyperdiploidy
- Gain of the odd numbered chromosomes including 3, 5, 7, 9, 11, 15, 19 and 21 is the other major abnormality observed in multiple myeloma.[5] [8]
- The mechanism of this gain phenomena is not well understood, one hypothesis being the gain of chromosome during single mitosis.[8]
| Chromosomal aberrations in multiple myeloma (MM) | ||
|---|---|---|
| Chromosomal Abnormality | Chromosome(s)/Protein(s) affected | Consequence |
| Trisomies | Odd-numbered chromosomes with the exception of chromosomes 1, 13, and 21 | |
| t(11;14)(q13;q32)
t(6;14q)(p21;32) t(12;14)(p13;q32) |
Cyclin D1
Cyclin D3 Cyclin D2 |
Over-expression; cell cycle dysregulation |
| t(4;14)(p16;q32) | FGFR3 or MMSET | Over-expression and activation; multiple myeloma cell proliferation/apoptosis prevention MMSET probably linked to crucial transforming event |
| t(14;16)(q32;q23)
t(14;20)(q32;q11) t(8;14)(q24;q32) |
c-MAF | Over-expression; involvement in IL-4 regulation |
| del 17p13 | p53 | Cell-cycle dysregulation/apoptosis |
| Monosomy 14 | Chromosome 14 | |
| Chromosome 13 deletion and monosomy | Chromosome 13 | |
| Gain(1q21) | Chromosome 1 | |
| Abbreviations used: FGFR3:fibroblast growth factor receptor 3; MMSET:multiple myeloma SET domain; MAF:musculoaponeurotic fibrosarcoma oncogene homolog. | ||
Mutations in myeloma
- Studies have demonstrated the presence of approximately 35 mutations per sample in multiple myeloma. These mutations cause loss of tumor suppressors and NFKB alterations.[8]
Tumor suppressors
- These mutations result in cell-cycle dysregulation with an increase in G1/S transition. Some of these mutations are mentioned in the table below.[8][44][45][46]
- The loss of tumor suppressors allows the cells to survive and grow without check points.
| Tumor suppressor genes commonly affected in myeloma |
|---|
| FAM46C (family with sequence similarity 46, member C) |
| DIS3 (Exosome complex exonuclease RRP44) |
| CYLD (Cylindromatosis) |
| Baculoviral IAP repeat containing protein 2 (BIRC2; also known as cIAP1) |
| BIRC3 (Baculoviral IAP repeat containing protein 3) |
| tumor necrosis factor receptor associated factor 3 (TRAF3) |
| CDKN2C |
| CDKN2A |
| TP53 |
NFKB alterations
- NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) is a protein complex which regulates DNA transcription, production of cytokine and cell survival. NF-κB plays a key role in generating cellular responses to stimuli such as stress, free radicals, heavy metals, ultraviolet radiations, and foreign antigens. NF-κB is also crucial in response of the immune system towards infections.[47][48][23]
- Deregulated NF-κB system leads to increased growth factors and cytokines such as IL-6 which promote growth and survival in myeloma cells.[23]
- Mutations in multiple genes can cause activation of NF-κB system which in turn plays a role in pathogenesis of multiple myeloma. Some of these mutations, that have been documented in genes regulating NF-κB pathway, are mentioned below.[49][50]
| Genes mutated associated with canonical signaling | Genes mutated associated with non-canonical signaling |
|---|---|
| TLR4
TNFRSF1A IKBKB IKBIP |
MAP3K14 |
Epigenetic changes in multiple myeloma
- Epigenetic events such as methylation of DNA and histones play an important role in pathogenesis of multiple myeloma.
- Of these events, the most notable is the global hypomethylation and gene-specific hypermethylation that takes place during progression of MGUS to myeloma.[8]
- A subset of patients with t(4;14) translocation show prominent DNA methylation changes that lead to MMSET overexpression.[8][39]
- Other dysregulations related to epigenetics in multiple myeloma may effect KDM6A (UTX), a histone demethylase, mixed lineage leukemia (MLL) protein, lysine demethylase 6B (KDM6B) and homeobox A9 (HOXA9).[8][51]
Bone marrow microenvironment and multiple myeloma
- Bone marrow microenvironment comprises of bone marrow stromal cells (BMSCs), extracellular matrix (ECM) proteins such as collagen, fibronectin and laminin, and the extracellular fluid containing cytokines and growth factors. This microenvironment is conducive to normal hematopoiesis but this also helps myeloma cells to have increased replication activity and anti-apoptotic resistance.[5][8][52]
- One of the key processes is the interaction and adherence of myeloma cells with bone marrow stromal cells (BMSCs) and extracellular matrix proteins through cellular adhesion molecules (CAMs) such as CD44 (H-CAM), CD56 (N-CAM), members of the CD49 integrin family (including very-late antigen VLA-4 and VLA-5), lymphocyte function-associated antigen-1, syndecan-1, and selectin.[5][8][53]
- This interaction leads to activation of p42/44 mitogen-activated protein kinase, and nuclear factor κB (NF-κB) which cause increased adhesion molecules, both on myeloma cells and bone marrow stromal cells. These events ultimately lead to increased production of cytokines. Of these cytokines IL-6, TNFα, BAFF, IGF and HGF are of particular importance.[5][8][54][55][56]
- These events cause localization of tumor cells in bone marrow, increased proliferation and survival, and resistance to apoptosis and chemotherapy. Bone marrow niche thus plays a crucial role in pathogenesis of multiple myeloma.[5][8][54][55][56]
- Decreased cellular adhesion molecules such as CD56 and chemokine receptors such as CXCR4 on myeloma cells lead to decreased adhesion to bone marrow stromal cells and immune evasion, allowing the myeloma cells to spread outside the bone marrow.[57][58]
Cytokines in multiple myeloma pathogenesis
- A variety of cytokines have been implicated in pathogenesis of multiple myeloma. Some of the relevant cytokines, their mechanisms and effects on tumor cells have been summarized in the table below.[5][59][60][60][61][62][63][64][65][66][67][68]
| Cytokines | Mechanism | Effects on tumor cells and pathogenesis |
|---|---|---|
| Interleukin 6 | Activates signal transduction pathways |
|
| Tumor necrosis factor α
TNF-α |
Activation of NF-κB
Activation of the MAPK pathways |
|
| B-cell activating factor
(BAFF) |
Activation of NF-κB |
|
| Insulin-like growth factor-1
(IGF-1) |
Activation of PI3K/Akt |
|
| Vascular endothelial growth factor
(VEGF) |
VEGF Receptor activation |
|
| Interleukin 17
(IL-17) |
Interleukin 17 receptors activation |
Pathophysiology of renal involvement
Abnormal antibody fragments are produced in multiple myeloma and are deposited in various organs, such as the kidneys. There are three major forms of renal damage in patients with multiple myeloma.
- Cast nephropathy: End-organ damage to the kidneys is typically due to light chain cast nephropathy. The pathophysiology of this type of renal involvement is based on light chain deposition in the renal tubules, which results in obstruction. Free light chains are readily filtered at the glomerulus and are reabsorbed in the proximal tubule of the nephron. This reabsorption occurs via the megalin-cubulin transport system.[69] In patients with multiple myeloma, there is excess production of free light chains, and the ability of the nephron to resorb light chains in the proximal tubule cannot meet the demands of the freely filtered light chains. This results in excess secretion of free light chains in the urine (known as Bence-Jones protein). Eosinophilic proteinaceous casts and crystalline structures can be seen. Cast formation occurs in the tubules due to excess abundance of free light chains that interact with Tamm-Horsfall proteins in the thick ascending loop of Henle.[69] Tubular obstruction ensues, triggering local inflammation which results in interstitial nephritis and fibrosis.[69] The onset of cast nephropathy can be very quick, requiring prompt treatment. Risk factors for development of cast nephropathy include monoclonal immunoglobulin secretion of >10 g/day, sepsis, and volume depletion.[70] Patients can also develop Fanconi syndrome, resulting in dysfunctional reabsorption ability by the proximal tubule, and type II renal tubular acidosis.
- Monoclonal immunoglobulin deposition disease (MIDD): In this subtype of renal involvement by multiple myeloma, the initial pathophysiological process is filtration of monoclonal immunoglobulins and subsequent deposition of immunoglobulins along the tubular or glomerular basement membrane.[70] Deposits of immunoglobulin can have a similar appearance as Kimmelstein-Wilson lesions (seen in diabetes). The immunoglobulins can appear fibroblast-like.
- Light chain amyloidosis: The pathophysiology of renal involvement by light chain amyloidosis begins with beta-pleated sheet formation in the tubules or glomeruli. Beta-pleated sheets form as a result of electrostatic interactions between heparan sulfate proteoglycan and amyloid proteins. Amyloid fibrils usually consist of immunoglobulin light chains (usually lambda light chain) and deposit in the basement membrane. The size of the fibrils vary from 7 to 10 nanometers. A diagnosis of this type of renal involvement is made by the visualization of apple green birefringence upon Congo red staining of the renal specimen.[70] It is frequently associated with nephrotic range proteinuria, in which greater than 3 grams of protein is excreted daily.
Pathophysiology of osseous involvement
Bone disease characterized by progressive osteolytic bone lesions leading to bone resorption is hallmark of multiple myeloma. Abnormal bone remodeling is thought to be the cause. It has been reported that up to 80% of the patients have characteristic osteolytic bone lesions at presentation and 60% of the patients with multiple myeloma will develop at least one pathological fracture at some stage.The pathophysiology of bony involvement in multiple myeloma is complex and is briefly described here..[71][72][73]
Increased osteoclastic activity
Osteoclasts are large, multinucleated cells of monocyte–macrophage lineage and play a crucial role in bone remodeling. Myeloma cells, in addition to their direct interaction with other cells, produce and release a number of factors which promote osteoclast differentiation and activation. Some of these factors and interactions have been described below in the tables.[71][74][75]
| Cell-cell interactions | Consequences |
|---|---|
| Myeloma cells to bone marrow stromal cells |
|
| Alpha4-beta1 integrin to vascular cell adhesion molecule 1 (VCAM-1) interaction |
|
| Myeloma cells to osteoblasts | Decreased production of OPG |
| Myeloma cells to osteoclasts | Direct adherence of myeloma cells to osteoclasts may result in
|
| Myeloma cells to immune cells | Increased production of cytokines, chemokines and factors associated with growth,
survival and migration. |
.
| Molecular pathways and factors associated with increased osteoclastic activity | Association with multiple myeloma |
|---|---|
| RANK/RANKL pathway
Binding of RANKL to RANK → fusion of osteoclast precursors into multinucleated cells → mature osteoclasts → ↑ bone resorption Osteoprotegerin (OPG) is a soluble decoy receptor for RANKL → ↓ binding of RANKL to RANK → ↓ bone resorption
|
Myeloma cells lead to increased RANKL and decreased OPG causing bone resorption and destruction. RANKL/OPG ratio is an idependent
prognostic factor in multiple myeloma
|
| Notch pathway
Binding of Notch receptors to its ligands → ↑ RANKL production → ↑ bone resorption
|
Myeloma cells express Notch 1,2,3 and their ligands Jagged 1,2 and Delta-like 1,3,4. This pathway, in addition to bone resorption, may also be involved in metastasis of myeloma cells by increasing expression of adhesion molecules, migratory chemokines, and angiogenetic factors, and disruption of the immune surveillance.
|
CCL-3 (MIP-1α)/CCL-20
|
|
| Activin A
Binds to type II transmembrane serine/threonine kinase receptor (ActRIIA/B) → recruitment and phosphorylation the type I receptor (ActRI, also called activin receptor-like kinase 4 (ALK4) receptor) → heterodimer formation → activation of the Smad signaling cascade → translocation of Smad2/3/4 complex in the nucleus → action as transcriptional factor → RANK expression and activation of NF-κB pathway → increased osteoclast differentiation |
|
Osteopontin
|
|
Interleukin 3 (IL-3)
|
|
Vascular endothelial growth factor (VEGF)
|
|
Interleukin 6 (IL-6)
|
|
Interleukin 17 (IL-17)
|
|
| B cell-activating factor (BAFF)
Binding to its receptor → activation of NF-κB → ↑ MM cell survival
|
|
| Bruton’s tyrosine kinase (BTK)
Osteoclast precursors with CXC chemokine receptor type 4 (CXCR4) and Bruton’s tyrosine kinase (BTK) expression → migration towards stromal cell-derived factor-1α (SDF-1α) → ↑ activation of Bruton’s tyrosine kinase (BTK) in myeloma cells.
|
|
| Stromal cell-derived factor-1α (SDF-1α)
Osteoclast precursors with CXC chemokine receptor type 4 (CXCR4) and Bruton’s tyrosine kinase (BTK) expression → migration towards stromal cell-derived factor-1α (SDF-1α) → ↑ activation of Bruton’s tyrosine kinase (BTK) in myeloma cells.
|
|
Annexin II (Annexin A2)
|
|
PU.1
|
Decreased osteoblast activity
Inhibition of osteoblasts leading to decreased bone formation is now considered to be a critical event in bone disease in multiple myeloma. This inhibition leads to bone loss as well as inability to repair the osteoytic lesions caused by increased osteoclastic activity. Several pathways and factors have been associated with suppressed osteoblastic activity. Some of them are mentioned below in the table.[71][74][75]
| Molecular pathways and factors associated with osteoblastic activity | Association with multiple myeloma |
|---|---|
Wingless and integration-1 (Wnt) signaling
Activation of Wnt pathway → binding of Wnt ligands to Wnt co-receptors LRP5/6 and one trans-membrane receptor of the FDZ family → formation of DVL–Axin–FRAT1–GSK-3β complex → translocation of β-catenin from cytoplasm to nucleus → activation of T cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors → ↑ bone formation and ↓ bone resorption Formation of WNT ligand–receptor tyrosine kinase-like orphan receptor 2 (ROR2) or the receptor-like tyrosine kinase (RYK)–FZD–DVL complex → activation of one of these 3 pathways: 1) Disheveled-associated activator of morphogenesis 1 (DAAM1)–RHO–RHO-associated kinase (ROCK) pathway; 2) RAC–Jun kinase (JNK)–RUNX2 pathway; WNT–Ca2+ pathway
|
Dickkopf 1 (Dkk-1)
Secreted frizzled related protein-2 (sFRP-2)
Runt-related transcription factor 2 (Runx2)/corebinding factor runt domain alpha subunit 1 (CBFA1)
|
| EphrinB2/EphB4 signaling pathway
Binding of ligands called ephrins (Eph receptor-interacting proteins) → activation of two cascades: 1) the forward signaling → ↑ osteoblast differentiation by downregulating RhoA; 2) the reverse signaling → ↓ osteoclast differentiation by ↓ Fos and Nfatc1 transcription
|
Multiple myeloma patients have decreased expression of both EphrinB2 and EphB4 in bone marrow stromal cells. |
Transforming growth factor-β (TGF-β)
|
Inhibition of Transforming Growth Factor-β have been shown to prevent myeloma cells to block osteoblastic differentiation. |
| Bone morphogenetic proteins (BMPs) | Myeloma cells express negative regulators of bone morphogenetic proteins that result in decreased activity of these proteins which in turn causes decreased osteoblastogenesis. |
Hepatocyte growth factor (HGF)
|
|
Interleukin 3 (IL-3)
|
Elevated levels had been demonstrated in bone marrow of myeloma patients. |
Interleukin 7 (IL-7)
|
|
Growth factor independence-1 (GFI1)
|
Bone marrow stromal cells (BMSCs) in myeloma patients have been shown to over-express growth factor independence-1 (GFI1). |
Tissue necrosis factor-α (TNF-α)
|
Tissue necrosis factor-α (TNF-α) levels are increased in multiple myeloma and it is thought to play a complex role in pathogenesis of multiple myeloma. |
| Adiponectin
adipocyte-derived hormone thought to act on osteoblasts and osteoclasts |
Increased adiponectin secretion through pharmacologic interventions have shown to decrease osteolytic lesions in myeloma. |
Renal involvement in multiple myeloma
Renal involvement is common in multiple myeloma as up to 50% of the patients go on to develop kidney disease at some point during the disease course. Half of these patients recover kidney function while the rest of them develop chronic kidney dysfunction. The causes of renal involvement in multiple myeloma are multiple and diverse. They can broadly be classified into Ig dependent and Ig independent mechanisms. Some of them have been discussed below in the table and then described briefly.[76][77][70]
| Ig-dependent renal injury | Ig-independent renal injury | ||
|---|---|---|---|
| Cause | Notes | Cause | Notes |
| Cast nephropathy (myeloma kidney) | Risk factors
|
Volume depletion | May cause
|
| Monoclonal Ig deposition disease | Sepsis | May cause
| |
| Light chain amyloidosis (AL) |
|
Hypercalcemia | May result in acute kidney injury directly or contribute to cast nephropathy |
| Glomerulonephritis | Following types have been demonstrated
|
Tumor lysis syndrome | Caused by uric acid or phosphate nephropathy |
| Tubulointerstitial nephritis | May be caused be either Ig-dependent or Ig-independent mechanisms | Direct parenchymal invasion by plasma cells | Rare cause. Association with advanced or aggressive multiple myeloma |
| Minimal change disease | Often with albuminuria and light chain proteinuria | Pyelonephritis | Rare cause. Pathogenesis is typically multifactorial and may include:
|
| Hyperviscosity syndrome | Seen primarily in cases of IgA, IgG3, or IgM myeloma | Medication toxicity | Following drugs may cause kidney injury:
|
| Henoch–Scholein purpura/IgA nephropathy | Associated with IgA myeloma | ||
| Immunotactoid glomerulopathy (and possibly fibrillary GN) | Rare conditions. The association between fibrillary disease and paraproteins is is not understood at this point. | ||
| Thrombotic microangiopathy (TMA) | Endothelial injury caused by paraprotein leads to thrombotic microangiopathy. | ||
| Membranous glomerulopathy | |||
Cast nephropathy (myeloma kidney)
- Normally circulating monoclonal free light chains are relatively freely filtered through the glomerulus and a receptor-mediated process leads to their endocytosis in proximal tubule cells. They bind to the tandem scavenger receptor system cubilin/megalin, followed by endocytosis through the clathrin-dependent endosomal/lysosomal pathway. They are then degraded within lysosomes.
- In multiple myeloma, myeloma cells produce excess of monoclonal free light chains that overwhelms the capacity of the proximal tubule cells to absorb and catabolize them. This leads to large amounts of monoclonal free light chains reaching distal renal tubules.
- In distal renal tubules, they interact with Tamm-Horsfall protein (uromodulin), a glycoprotein produced in the medullary thick ascending limb of the loop of Henle in kidneys. This interaction leads to formation of myeloma casts.
- These casts block the glomerular flow, compromising renal function and may lead to proximal tubular atrophy. This may also contribute to interstitial fibrosis.
- This blockage leads to increased luminal pressure, resulting in decreased blood flow and causing further renal injury.
- The excessive amounts of monoclonal free light chains being absorbed in proximal renal tubules leads to induction of apoptosis and DNA damage in proximal renal tubules.
- Free light chains also leads to increased pro-inflammatory cytokines such as IL-6, macrophage chemoattractant protein-1 (MCP-1), and tumor necrosis factor α (TNFα) through transcription factors nuclear factor kappa B (NFκB) and mitogen-activated protein kinases.
- All these events leads to kidney injury and inability of renal cells to repair the injury. On histopathological studies, chronic tubulointerstitial nephropathy with marked tubular atrophy, laminated casts in the tubules, and interstitial fibrosis may be observed.
| Factors associated with increased risk
of AKI or CKD in multiple myeloma |
Mechanism of injury |
|---|---|
| Volume depletion | Increased FLC concentration
Decreased GFR Pre-renal azotemia |
| Hypercalcemia | Renal vasoconstriction
Decreased GFR |
| Iodinated contrast media | Nephrotoxicity |
| Nonsteroidal anti-inflammatory drugs | Renal vasoconstriction
Decreased GFR |
| Diuretics | Increased sodium chloride concentration
Increased cast formation |
| Aminoglycosides | Renal vasoconstriction
Decreased GFR |
| Hyperuricemia | |
| Comorbidities such as chronic kidney disease, diabetes, aging, hypertension, and cardiovascular disease. | |
Hypercalcemia
- Hypercalcemia in multiple myeloma primarily results from increased osteoclastic bone resorption. It is the second most common cause of renal injury in myeloma patients.
- Hypercalcemia leads to intra-tubular calcium deposition and vasoconstriction in renal vasculature. The resultant decrease in GFR not only causes renal injury by itself but also favors cast formation.
- By impairing renal concentrating ability through resistance to anti-diuretic hormone, it may also cause nephrogenic diabetes insipidus. Polyuria may develop, leading to hypovolemia. Net effect is decreased GFR, concentrated urine, reduced urine flow, and pre-renal azotemia.
Light-chain glomerulopathy
- Deposition of immunoglobulins either in the form of amyloid or non-amyloid, typically resulting in non-selective proteinuria.
- Amyloid deposits are fibrillar structures, consist of variable region fragments of the light chain, and are seen primarily within the glomeruli.
- Some patients may have predominant vascular amyloid deposits rather than glomerular deposits and present with renal failure instead of nephrotic syndrome.
Light chain deposition disease (LCDD)
- Non-fibrillar, granular deposits are topically observed in mesangial area along with thickening of basement membrane which resembles type II membranoproliferative glomerulonephritis or diabetic glomerulopathy. The light chain deposits may also be observed in renal vasculature in addition to mesangial area.
- Typical clinical picture is that of nephrotic syndrome but renal impairment is more severe with almost all patients developing renal failure.
Immune dysfunction in multiple myeloma
- Immunodeficiency is an important characteristic of multiple myeloma and is thought to associated with increased risk for infections, and affects disease progression and response to treatment.[5][78]
- The typical clinical picture is that of global immunosuppression. The interaction between myeloma cells and bone marrow stromal cells and cytokines produced as a result of this interaction are thought to play a key role.[5][78][79][80]
- Following abnormalities have been demonstrated in myeloma patients:[79][81][80]
- Decreased numbers of natural killer cells, B-cells, and memory T cells
- Decreased non-myeloma immunoglobulins
- Decreased expression of cell-surface markers on CD4+ and CD8+ T cells such as CD28 and CD152. These markers are associated with cell-signaling.
- Abnormal signal transduction in T-cells
- Abnormalities in cytokine production
- Abnormalities in antibodies response and production
- Aberrations in B-cells maturation
- Defects in natural killer cells and antigen presenting cells
Gross Pathology
-
Vertebrae in multiple myeloma
(Image courtesy of Melih Aktan M.D.) -
Calvarium in multiple myeloma.
(Image courtesy of Melih Aktan M.D.)
Microscopic Pathology
On microscopic histopathological analysis, multiple myeloma is characterized by the following:[4]
- Abundant eosinophilic cytoplasm
- Eccentrically placed nucleus
- Clock face morphology of the nucleus due to chromatin clumps around the edges
- Russell bodies which are eosinophilic, large (10-15 micrometres), homogenous immunoglobulin-containing inclusions
- Dutcher bodies which are PAS stain +ve intranuclear crystalline rods
- Shown below is a series of microscopic images seen in multiple myeloma:
-
Bone marrow aspiration in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow biopsy in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Bone marrow in multiple myeloma.
(Image courtesy of Melih Aktan M.D.) -
Multiple myeloma slide with intermediate magnification[4]
-
Multiple myeloma slide with high magnification[4]
-
Multiple myeloma slide with russell bodies[4]
References
- ↑ Multiple myeloma. Radiopaedia (2015)http://radiopaedia.org/articles/multiple-myeloma-1 Accessed on September, 20th 2015
- ↑ Multiple myeloma. Wikipedia (2015)https://en.wikipedia.org/wiki/Multiple_myeloma#Pathophysiology Accessed on September, 20th 2015
- ↑ Multiple myeloma. Medlineplus (2015)https://www.nlm.nih.gov/medlineplus/multiplemyeloma.html Accessed on September, 20th 2015
- ↑ 4.0 4.1 4.2 4.3 4.4 Multiple myeloma. Librepathology (2015)http://www.wikidoc.org/index.php?title=Multiple_myeloma_pathophysiology&action=edit§ion=1 Accessed on September, 20th 2015
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 Borrello I (November 2012). "Can we change the disease biology of multiple myeloma?". Leuk. Res. 36 Suppl 1: S3–12. doi:10.1016/S0145-2126(12)70003-6. PMC 3698609. PMID 23176722.
- ↑ 6.0 6.1 6.2 6.3 Chng WJ, Glebov O, Bergsagel PL, Kuehl WM (December 2007). "Genetic events in the pathogenesis of multiple myeloma". Best Pract Res Clin Haematol. 20 (4): 571–96. doi:10.1016/j.beha.2007.08.004. PMC 2198931. PMID 18070707.
- ↑ 7.0 7.1 7.2 7.3 Hengeveld PJ, Kersten MJ (February 2015). "B-cell activating factor in the pathophysiology of multiple myeloma: a target for therapy?". Blood Cancer J. 5: e282. doi:10.1038/bcj.2015.3. PMC 4349256. PMID 25723853.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 8.14 8.15 8.16 8.17 Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (July 2001). "Plasma cell differentiation requires the transcription factor XBP-1". Nature. 412 (6844): 300–7. doi:10.1038/35085509. PMID 11460154.
- ↑ Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (October 2011). "The genetic network controlling plasma cell differentiation". Semin. Immunol. 23 (5): 341–9. doi:10.1016/j.smim.2011.08.010. PMID 21924923.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (July 2001). "Plasma cell differentiation requires the transcription factor XBP-1". Nature. 412 (6844): 300–7. doi:10.1038/35085509. PMID 11460154.
- ↑ Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (October 2011). "The genetic network controlling plasma cell differentiation". Semin. Immunol. 23 (5): 341–9. doi:10.1016/j.smim.2011.08.010. PMID 21924923.
- ↑ 14.0 14.1 Eleonora Market, F. Nina Papavasiliou (2003) V(D)J Recombination and the Evolution of the Adaptive Immune System PLoS Biology1(1): e16.
- ↑ 15.0 15.1 Litman GW, Rast JP, Shamblott MJ; et al. (1993). "Phylogenetic diversification of immunoglobulin genes and the antibody repertoire". Mol. Biol. Evol. 10 (1): 60–72. PMID 8450761.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (October 2011). "The genetic network controlling plasma cell differentiation". Semin. Immunol. 23 (5): 341–9. doi:10.1016/j.smim.2011.08.010. PMID 21924923.
- ↑ Boyle EM, Davies FE, Leleu X, Morgan GJ (April 2014). "Understanding the multiple biological aspects leading to myeloma". Haematologica. 99 (4): 605–12. doi:10.3324/haematol.2013.097907. PMC 3971069. PMID 24688108.
- ↑ Keim C, Kazadi D, Rothschild G, Basu U (January 2013). "Regulation of AID, the B-cell genome mutator". Genes Dev. 27 (1): 1–17. doi:10.1101/gad.200014.112. PMC 3553278. PMID 23307864.
- ↑ González D, van der Burg M, García-Sanz R, Fenton JA, Langerak AW, González M, van Dongen JJ, San Miguel JF, Morgan GJ (November 2007). "Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma". Blood. 110 (9): 3112–21. doi:10.1182/blood-2007-02-069625. PMID 17634408.
- ↑
- ↑ 22.0 22.1 22.2 22.3 Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ (March 2004). "Characterization of clonogenic multiple myeloma cells". Blood. 103 (6): 2332–6. doi:10.1182/blood-2003-09-3064. PMC 3311914. PMID 14630803.
- ↑ 23.0 23.1 23.2 23.3 Tian B, Brasier AR (2003). "Identification of a nuclear factor kappa B-dependent gene network". Recent Prog. Horm. Res. 58: 95–130. PMID 12795416.
- ↑ Huff CA, Matsui W (June 2008). "Multiple myeloma cancer stem cells". J. Clin. Oncol. 26 (17): 2895–900. doi:10.1200/JCO.2007.15.8428. PMC 2610256. PMID 18539970.
- ↑ Bataille R, Jégo G, Robillard N, Barillé-Nion S, Harousseau JL, Moreau P, Amiot M, Pellat-Deceunynck C (September 2006). "The phenotype of normal, reactive and malignant plasma cells. Identification of "many and multiple myelomas" and of new targets for myeloma therapy". Haematologica. 91 (9): 1234–40. PMID 16956823.
- ↑ Grogan TM, Durie BG, Lomen C, Spier C, Wirt DP, Nagle R, Wilson GS, Richter L, Vela E, Maxey V (October 1987). "Delineation of a novel pre-B cell component in plasma cell myeloma: immunochemical, immunophenotypic, genotypic, cytologic, cell culture, and kinetic features". Blood. 70 (4): 932–42. PMID 3115338.
- ↑ Kiel K, Cremer FW, Rottenburger C, Kallmeyer C, Ehrbrecht E, Atzberger A, Hegenbart U, Goldschmidt H, Moos M (May 1999). "Analysis of circulating tumor cells in patients with multiple myeloma during the course of high-dose therapy with peripheral blood stem cell transplantation". Bone Marrow Transplant. 23 (10): 1019–27. doi:10.1038/sj.bmt.1701767. PMID 10373068.
- ↑ Morgan GJ, Davies FE, Linet M (July 2002). "Myeloma aetiology and epidemiology". Biomed. Pharmacother. 56 (5): 223–34. PMID 12199621.
- ↑ Wadhera RK, Rajkumar SV (October 2010). "Prevalence of monoclonal gammopathy of undetermined significance: a systematic review". Mayo Clin. Proc. 85 (10): 933–42. doi:10.4065/mcp.2010.0337. PMC 2947966. PMID 20713974.
- ↑ Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D, Hoover R, Rajkumar SV (May 2009). "Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study". Blood. 113 (22): 5412–7. doi:10.1182/blood-2008-12-194241. PMC 2689042. PMID 19179464.
- ↑ Wang SS, Voutsinas J, Chang ET, Clarke CA, Lu Y, Ma H, West D, Lacey JV, Bernstein L (July 2013). "Anthropometric, behavioral, and female reproductive factors and risk of multiple myeloma: a pooled analysis". Cancer Causes Control. 24 (7): 1279–89. doi:10.1007/s10552-013-0206-0. PMC 3684420. PMID 23568533.
- ↑ Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM (July 2007). "Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis". Lancet. 370 (9581): 59–67. doi:10.1016/S0140-6736(07)61050-2. PMID 17617273.
- ↑ Kachuri L, Demers PA, Blair A, Spinelli JJ, Pahwa M, McLaughlin JR, Pahwa P, Dosman JA, Harris SA (October 2013). "Multiple pesticide exposures and the risk of multiple myeloma in Canadian men". Int. J. Cancer. 133 (8): 1846–58. doi:10.1002/ijc.28191. PMID 23564249.
- ↑ Singh J, Dudley AW, Kulig KA (December 1990). "Increased incidence of monoclonal gammopathy of undetermined significance in blacks and its age-related differences with whites on the basis of a study of 397 men and one woman in a hospital setting". J. Lab. Clin. Med. 116 (6): 785–9. PMID 2246554.
- ↑ Kristinsson SY, Björkholm M, Goldin LR, Blimark C, Mellqvist UH, Wahlin A, Turesson I, Landgren O (November 2009). "Patterns of hematologic malignancies and solid tumors among 37,838 first-degree relatives of 13,896 patients with multiple myeloma in Sweden". Int. J. Cancer. 125 (9): 2147–50. doi:10.1002/ijc.24514. PMC 2737604. PMID 19582882.
- ↑ Broderick P, Chubb D, Johnson DC, Weinhold N, Försti A, Lloyd A, Olver B, Ma Y, Dobbins SE, Walker BA, Davies FE, Gregory WA, Childs JA, Ross FM, Jackson GH, Neben K, Jauch A, Hoffmann P, Mühleisen TW, Nöthen MM, Moebus S, Tomlinson IP, Goldschmidt H, Hemminki K, Morgan GJ, Houlston RS (November 2011). "Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk". Nat. Genet. 44 (1): 58–61. doi:10.1038/ng.993. PMC 5108406. PMID 22120009.
- ↑ Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Försti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, Hose D, Walker BA, Davies FE, Gregory WA, Jackson GH, Irving JA, Pratt G, Fegan C, Fenton JA, Neben K, Hoffmann P, Nöthen MM, Mühleisen TW, Eisele L, Ross FM, Straka C, Einsele H, Langer C, Dörner E, Allan JM, Jauch A, Morgan GJ, Hemminki K, Houlston RS, Goldschmidt H (October 2013). "Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk". Nat. Genet. 45 (10): 1221–1225. doi:10.1038/ng.2733. PMC 5053356. PMID 23955597.
- ↑ Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, Ross FM, Davies FE, Gonzalez D, Morgan GJ (April 2013). "Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells". Blood. 121 (17): 3413–9. doi:10.1182/blood-2012-12-471888. PMID 23435460.
- ↑ 39.0 39.1 Chesi M, Nardini E, Brents LA, Schröck E, Ried T, Kuehl WM, Bergsagel PL (July 1997). "Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3". Nat. Genet. 16 (3): 260–4. doi:10.1038/ng0797-260. PMC 3901950. PMID 9207791.
- ↑ Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H (December 2009). "International Myeloma Working Group molecular classification of multiple myeloma: spotlight review". Leukemia. 23 (12): 2210–21. doi:10.1038/leu.2009.174. PMC 2964268. PMID 19798094.
- ↑ Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, Avramidou A, Irving JA, Gonzalez D, Davies FE, Morgan GJ (January 2009). "MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells". Haematologica. 94 (1): 78–86. doi:10.3324/haematol.13426. PMC 2625417. PMID 19059936.
- ↑ Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM (January 2000). "Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma". Proc. Natl. Acad. Sci. U.S.A. 97 (1): 228–33. PMC 26645. PMID 10618400.
- ↑ Nobuyoshi M, Kawano M, Tanaka H, Ishikawa H, Tanabe O, Iwato K, Asaoku H, Sakai A, Kuramoto A (April 1991). "Increased expression of the c-myc gene may be related to the aggressive transformation of human myeloma cells". Br. J. Haematol. 77 (4): 523–8. PMID 2025578.
- ↑ Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, Johnson DC, Gonzalez D, Dagrada GP, Protheroe RK, Konn ZJ, Stockley DM, Gregory WM, Davies FE, Ross FM, Morgan GJ (October 2010). "A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value". Blood. 116 (15): e56–65. doi:10.1182/blood-2010-04-279596. PMID 20616218.
- ↑ Gonzalez-Paz N, Chng WJ, McClure RF, Blood E, Oken MM, Van Ness B, James CD, Kurtin PJ, Henderson K, Ahmann GJ, Gertz M, Lacy M, Dispenzieri A, Greipp PR, Fonseca R (February 2007). "Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications". Blood. 109 (3): 1228–32. doi:10.1182/blood-2006-05-024661. PMID 16840723.
- ↑ Seemann S, Maurici D, Olivier M, Caron de Fromentel C, Hainaut P (2004). "The tumor suppressor gene TP53: implications for cancer management and therapy". Crit Rev Clin Lab Sci. 41 (5–6): 551–83. doi:10.1080/10408360490504952. PMID 15603511.
- ↑ Gilmore TD (October 2006). "Introduction to NF-kappaB: players, pathways, perspectives". Oncogene. 25 (51): 6680–4. doi:10.1038/sj.onc.1209954. PMID 17072321.
- ↑ Perkins ND (January 2007). "Integrating cell-signalling pathways with NF-kappaB and IKK function". Nat. Rev. Mol. Cell Biol. 8 (1): 49–62. doi:10.1038/nrm2083. PMID 17183360.
- ↑ Roy P, Sarkar UA, Basak S (May 2018). "The NF-κB Activating Pathways in Multiple Myeloma". Biomedicines. 6 (2). doi:10.3390/biomedicines6020059. PMC 6027071. PMID 29772694.
- ↑ Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR (March 2011). "Initial genome sequencing and analysis of multiple myeloma". Nature. 471 (7339): 467–72. doi:10.1038/nature09837. PMC 3560292. PMID 21430775.
- ↑ van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA (May 2009). "Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer". Nat. Genet. 41 (5): 521–3. doi:10.1038/ng.349. PMC 2873835. PMID 19330029.
- ↑ Mitsiades CS, Mitsiades NS, Munshi NC, Richardson PG, Anderson KC (July 2006). "The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions". Eur. J. Cancer. 42 (11): 1564–73. doi:10.1016/j.ejca.2005.12.025. PMID 16765041.
- ↑ Teoh G, Anderson KC (February 1997). "Interaction of tumor and host cells with adhesion and extracellular matrix molecules in the development of multiple myeloma". Hematol. Oncol. Clin. North Am. 11 (1): 27–42. PMID 9081202.
- ↑ 54.0 54.1 Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA, Anderson KC (February 1996). "Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B". Blood. 87 (3): 1104–12. PMID 8562936.
- ↑ 55.0 55.1 Moreaux J, Legouffe E, Jourdan E, Quittet P, Rème T, Lugagne C, Moine P, Rossi JF, Klein B, Tarte K (April 2004). "BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone". Blood. 103 (8): 3148–57. doi:10.1182/blood-2003-06-1984. PMC 2387243. PMID 15070697.
- ↑ 56.0 56.1 Derksen PW, Keehnen RM, Evers LM, van Oers MH, Spaargaren M, Pals ST (February 2002). "Cell surface proteoglycan syndecan-1 mediates hepatocyte growth factor binding and promotes Met signaling in multiple myeloma". Blood. 99 (4): 1405–10. PMID 11830493.
- ↑ Pellat-Deceunynck C, Barillé S, Jego G, Puthier D, Robillard N, Pineau D, Rapp MJ, Harousseau JL, Amiot M, Bataille R (December 1998). "The absence of CD56 (NCAM) on malignant plasma cells is a hallmark of plasma cell leukemia and of a special subset of multiple myeloma". Leukemia. 12 (12): 1977–82. PMID 9844928.
- ↑ Barker HF, Hamilton MS, Ball J, Drew M, Franklin IM (July 1992). "Expression of adhesion molecules LFA-3 and N-CAM on normal and malignant human plasma cells". Br. J. Haematol. 81 (3): 331–5. PMID 1382543.
- ↑ Seidl S, Kaufmann H, Drach J (September 2003). "New insights into the pathophysiology of multiple myeloma". Lancet Oncol. 4 (9): 557–64. PMID 12965277.
- ↑ 60.0 60.1 Gupta D, Treon SP, Shima Y, Hideshima T, Podar K, Tai YT, Lin B, Lentzsch S, Davies FE, Chauhan D, Schlossman RL, Richardson P, Ralph P, Wu L, Payvandi F, Muller G, Stirling DI, Anderson KC (December 2001). "Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications". Leukemia. 15 (12): 1950–61. PMID 11753617.
- ↑ Chauhan D, Kharbanda S, Ogata A, Urashima M, Teoh G, Robertson M, Kufe DW, Anderson KC (January 1997). "Interleukin-6 inhibits Fas-induced apoptosis and stress-activated protein kinase activation in multiple myeloma cells". Blood. 89 (1): 227–34. PMID 8978296.
- ↑ Hideshima T, Chauhan D, Schlossman R, Richardson P, Anderson KC (July 2001). "The role of tumor necrosis factor alpha in the pathophysiology of human multiple myeloma: therapeutic applications". Oncogene. 20 (33): 4519–27. doi:10.1038/sj.onc.1204623. PMID 11494147.
- ↑ Jourdan M, Tarte K, Legouffe E, Brochier J, Rossi JF, Klein B (March 1999). "Tumor necrosis factor is a survival and proliferation factor for human myeloma cells". Eur. Cytokine Netw. 10 (1): 65–70. PMC 2025696. PMID 10210775.
- ↑ Tai YT, Li XF, Breitkreutz I, Song W, Neri P, Catley L, Podar K, Hideshima T, Chauhan D, Raje N, Schlossman R, Richardson P, Munshi NC, Anderson KC (July 2006). "Role of B-cell-activating factor in adhesion and growth of human multiple myeloma cells in the bone marrow microenvironment". Cancer Res. 66 (13): 6675–82. doi:10.1158/0008-5472.CAN-06-0190. PMID 16818641.
- ↑ Georgii-Hemming P, Wiklund HJ, Ljunggren O, Nilsson K (September 1996). "Insulin-like growth factor I is a growth and survival factor in human multiple myeloma cell lines". Blood. 88 (6): 2250–8. PMID 8822946.
- ↑ Dankbar B, Padró T, Leo R, Feldmann B, Kropff M, Mesters RM, Serve H, Berdel WE, Kienast J (April 2000). "Vascular endothelial growth factor and interleukin-6 in paracrine tumor-stromal cell interactions in multiple myeloma". Blood. 95 (8): 2630–6. PMID 10753844.
- ↑ Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, Pai C, Amin S, Tai YT, Richardson PG, Ghobrial IM, Treon SP, Daley JF, Anderson KC, Kutok JL, Munshi NC (July 2010). "Elevated IL-17 produced by TH17 cells promotes myeloma cell growth and inhibits immune function in multiple myeloma". Blood. 115 (26): 5385–92. doi:10.1182/blood-2009-10-246660. PMC 2902136. PMID 20395418.
- ↑ Noonan K, Marchionni L, Anderson J, Pardoll D, Roodman GD, Borrello I (November 2010). "A novel role of IL-17-producing lymphocytes in mediating lytic bone disease in multiple myeloma". Blood. 116 (18): 3554–63. doi:10.1182/blood-2010-05-283895. PMC 4017298. PMID 20664052.
- ↑ 69.0 69.1 69.2 Finkel KW, Cohen EP, Shirali A, Abudayyeh A, American Society of Nephrology Onco-Nephrology Forum (2016). "Paraprotein-Related Kidney Disease: Evaluation and Treatment of Myeloma Cast Nephropathy". Clin J Am Soc Nephrol. 11 (12): 2273–2279. doi:10.2215/CJN.01640216. PMC 5142056. PMID 27526708.
- ↑ 70.0 70.1 70.2 70.3 Heher EC, Rennke HG, Laubach JP, Richardson PG (2013). "Kidney disease and multiple myeloma". Clin J Am Soc Nephrol. 8 (11): 2007–17. doi:10.2215/CJN.12231212. PMC 3817918. PMID 23868898.
- ↑ 71.0 71.1 71.2 Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA (January 2018). "Pathogenesis of bone disease in multiple myeloma: from bench to bedside". Blood Cancer J. 8 (1): 7. doi:10.1038/s41408-017-0037-4. PMC 5802524. PMID 29330358.
- ↑ Terpos E, Morgan G, Dimopoulos MA, Drake MT, Lentzsch S, Raje N, Sezer O, García-Sanz R, Shimizu K, Turesson I, Reiman T, Jurczyszyn A, Merlini G, Spencer A, Leleu X, Cavo M, Munshi N, Rajkumar SV, Durie BG, Roodman GD (June 2013). "International Myeloma Working Group recommendations for the treatment of multiple myeloma-related bone disease". J. Clin. Oncol. 31 (18): 2347–57. doi:10.1200/JCO.2012.47.7901. PMC 4878084. PMID 23690408.
- ↑ Terpos E, Berenson J, Cook RJ, Lipton A, Coleman RE (May 2010). "Prognostic variables for survival and skeletal complications in patients with multiple myeloma osteolytic bone disease". Leukemia. 24 (5): 1043–9. doi:10.1038/leu.2010.62. PMID 20376081.
- ↑ 74.0 74.1 Edwards CM, Zhuang J, Mundy GR (June 2008). "The pathogenesis of the bone disease of multiple myeloma". Bone. 42 (6): 1007–13. doi:10.1016/j.bone.2008.01.027. PMC 2474770. PMID 18406675.
- ↑ 75.0 75.1 Hameed A, Brady JJ, Dowling P, Clynes M, O'Gorman P (2014). "Bone disease in multiple myeloma: pathophysiology and management". Cancer Growth Metastasis. 7: 33–42. doi:10.4137/CGM.S16817. PMC 4133035. PMID 25187738.
- ↑ Dimopoulos MA, Kastritis E, Rosinol L, Bladé J, Ludwig H (August 2008). "Pathogenesis and treatment of renal failure in multiple myeloma". Leukemia. 22 (8): 1485–93. doi:10.1038/leu.2008.131. PMID 18528426.
- ↑ Mussap M, Merlini G (2014). "Pathogenesis of renal failure in multiple myeloma: any role of contrast media?". Biomed Res Int. 2014: 167125. doi:10.1155/2014/167125. PMC 4022292. PMID 24877060.
- ↑ 78.0 78.1 Cook G, Campbell JD (September 1999). "Immune regulation in multiple myeloma: the host-tumour conflict". Blood Rev. 13 (3): 151–62. doi:10.1054/blre.1999.0111.
- ↑ 79.0 79.1 Schütt P, Brandhorst D, Stellberg W, Poser M, Ebeling P, Müller S, Buttkereit U, Opalka B, Lindemann M, Grosse-Wilde H, Seeber S, Moritz T, Nowrousian MR (August 2006). "Immune parameters in multiple myeloma patients: influence of treatment and correlation with opportunistic infections". Leuk. Lymphoma. 47 (8): 1570–82. doi:10.1080/10428190500472503. PMID 16966269.
- ↑ 80.0 80.1 Urashima M, Ogata A, Chauhan D, Hatziyanni M, Vidriales MB, Dedera DA, Schlossman RL, Anderson KC (March 1996). "Transforming growth factor-beta1: differential effects on multiple myeloma versus normal B cells". Blood. 87 (5): 1928–38. PMID 8634441.
- ↑ Mozaffari F, Hansson L, Kiaii S, Ju X, Rossmann ED, Rabbani H, Mellstedt H, Osterborg A (February 2004). "Signalling molecules and cytokine production in T cells of multiple myeloma-increased abnormalities with advancing stage". Br. J. Haematol. 124 (3): 315–24. PMID 14717778.
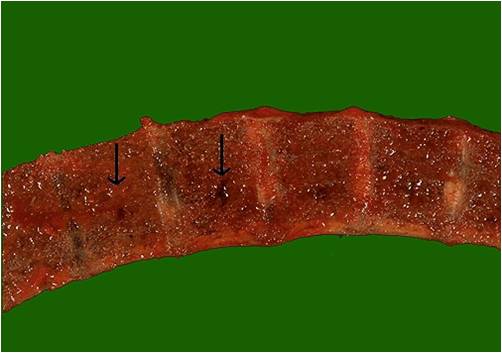
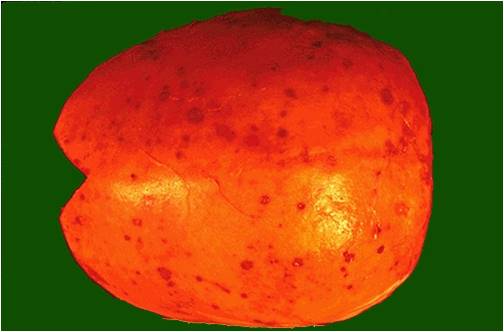
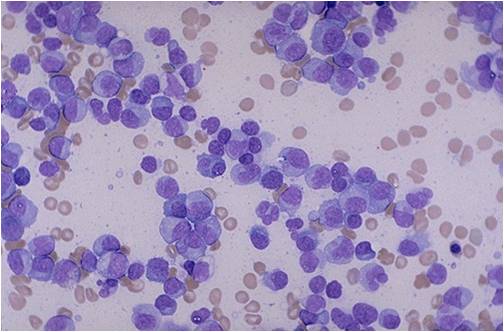
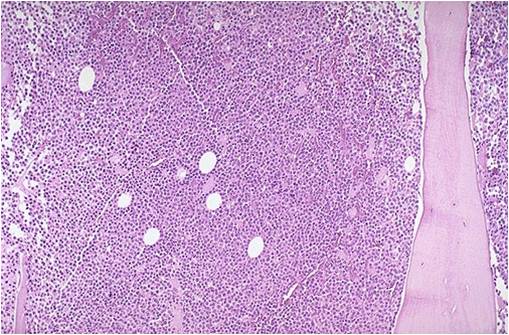

![Multiple myeloma slide with intermediate magnification[4]](/images/4/40/Multiple_myeloma_intermed_mag.jpg)
![Multiple myeloma slide with high magnification[4]](/images/d/d5/Multiple_myeloma.jpg)
![Multiple myeloma slide with russell bodies[4]](/images/a/ad/Russell_bodies.jpg)