TATA box gene transcriptions
Editor-In-Chief: Henry A. Hoff
The TATA box (also called Goldberg-Hogness box)[1] is a DNA sequence (cis-regulatory element) found in the promoter region of genes in archaea and eukaryotes;[2] approximately 24% of human genes contain a TATA box within the core promoter.[3]
The TATA box is a binding site of either general transcription factors or histones.
Consensus sequences
In the direction of transcription along the DNA strand, the TATA box has the core DNA sequence 3'-TATAAA-5' or a variant, which is usually followed by three or more adenine (A) bases, specifically [3'-TATAAA(A)AAA-5' on the template strand].
"[M]ost of the diversity within metazoan core promoters appears to involve the variable occurrence of consensus or near-consensus TATA, Inr, and DPE elements."[4]
The TATA box can be an AT-rich sequence "located at a fixed distance upstream of the transcription start site"[2].
Histones
The binding of a transcription factor blocks the binding of a histone and vice versa.
Gene expressions
Although it is harder to regulate the transcription of genes with multiple transcription start sites, "variations in the expression of a constitutive gene would be minimized by the use of multiple start sites."[5]
Earlier "studies led to the design of a super core promoter (SCP) that contains a TATA, Inr, MTE, and DPE in a single promoter (Juven-Gershon et al., 2006b). The SCP is the strongest core promoter observed in vitro and in cultured cells and yields high levels of transcription in conjunction with transcriptional enhancers. These findings indicate that gene expression levels can be modulated via the core promoter."[5]
Gene ID: 60
The "natural serum-responsive upstream activating sequence (UAS) of the human β-actin gene (ACTB) selectively activates TATA box-dependent but not INR-dependent transcription via a mechanism that involves the serum response factor (SRF) activator and the TATA-binding/bending activity of TBP in live human cells."[6]
Human genes
"TATA-containing genes are more often highly regulated, such as by biotic or stress stimuli."[3] Only "∼10% of these TATA-containing promoters have the canonical TATA box (TATAWAWR)."[3]
"SRF-regulated genes of the actin/cytoskeleton/contractile family tend to have a TATA box."[6]
Different "TATA box sequences have different abilities to convey the activating signals of certain enhancers and activators in mammalian cells [...] and in yeast [...]."[6]
"SRF is a well established master regulator of the specific family of genes encoding the actin cytoskeleton and contractile apparatus [...], and we found that ~40% of the core promoters for these genes contain a TATA box, which is a significant enrichment compared to the low overall frequency of TATA-containing promoters in human and mouse genomes (...)."[6] "Global frequencies of core promoter types for human [9010 orthologous mouse-human promoter pairs with 1848 TATA-containing or 7162 TATA-less][7] genes with experimentally validated transcription start sites [are known from 2006]."[6] "The TATA box [...] has a consensus sequence of TATAWAAR [...]."[6] W = A or T and R = A or G. We "estimate that ~17% of promoters contain a TATA box".[7]
Gene transcriptions
"From a teleological standpoint, this arrangement [of focused promoters] is consistent with the notion that it would be easier to regulate the transcription of a gene with a single transcription start site than one with multiple start sites."[5]
The TATA box is involved in the process of transcription by RNA polymerase.
Approximately “76% of human core promoters lack TATA-like elements, have a high GC content, and are enriched in Sp1 binding sites.”[3]
"[T]wo motifs - M3 (SCGGAAGY) and M22 (TGCGCANK) - ... occur preferentially in human TATA-less core promoters."[3]
"About 24% of human genes have a TATA-like element and their promoters are generally AT-rich; however, only ~10% of these TATA-containing promoters have the canonical TATA box (TATAWAWR). In contrast, ~46% of human core promoters contain the consensus INR (YYANWYY) and ~30% are INR-containing TATA-less genes."[3] W = A or T, Y = C or T, N = G, A, T, or C, and R = A or G.
Apparently, another ~46% of human promoters lack both TATA-like and consensus INR elements.
Transcription start sites
The consensus sequence is usually located 25 base pairs [(bps) or nucleotides (nts)] upstream [(-)] of the transcription site; i.e., the transcription start site (TSS).
Focused promoters
"In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms."[5]
"Focused transcription initiation occurs in all organisms, and appears to be the predominant or exclusive mode of transcription in simpler organisms."[5]
"In vertebrates, focused transcription tends to be associated with regulated promoters".[5]
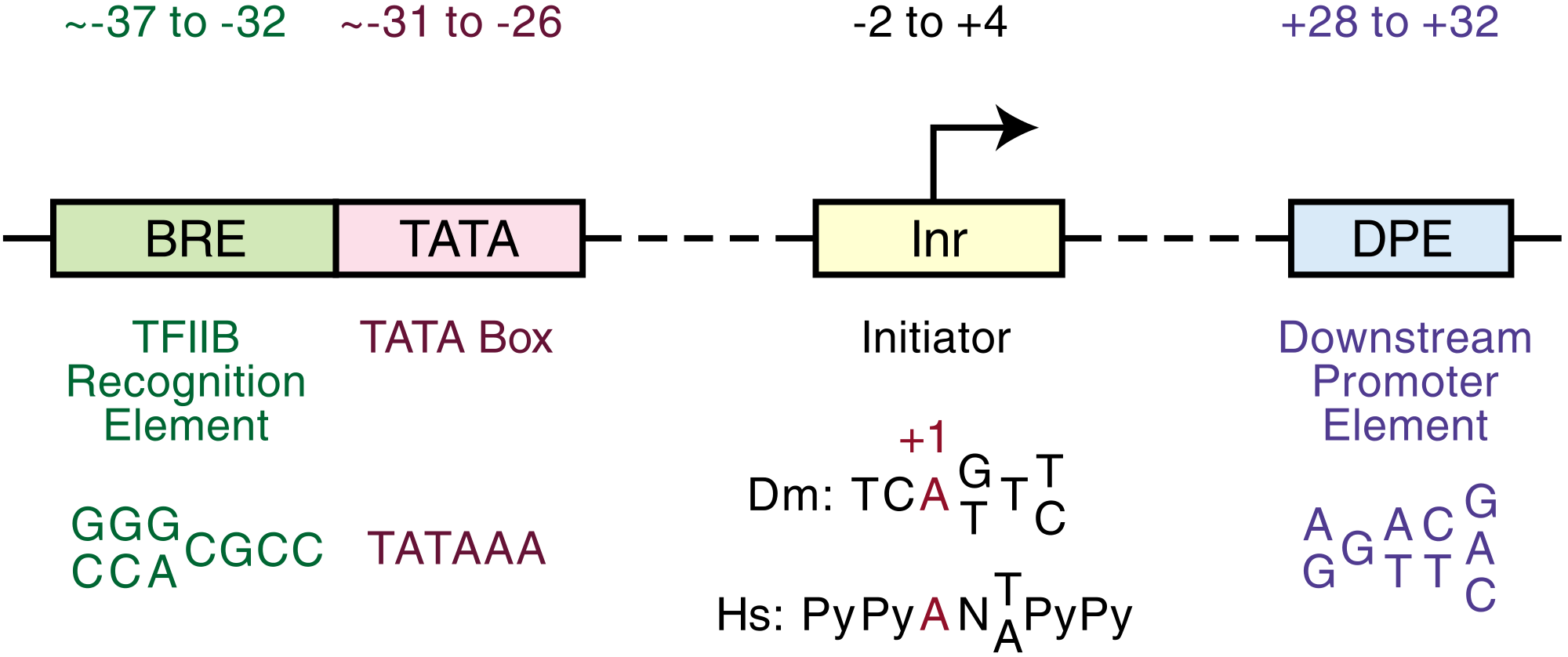
"The analysis of focused core promoters has led to the discovery of sequence motifs such as the TATA box, BREu (upstream TFIIBrecognition element), Inr (initiator), MTE (motif ten element), DPE (downstream promoter element), DCE (downstream core element), and XCPE1 (Xcore promoter element 1) [...]."[5]
Dispersed promoters
"In dispersed transcription, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes."[5]
In vertebrates, "dispersed transcription is typically observed in constitutive promoters in CpG islands."[5]
Core promoters
"Focused transcription typically initiates within the Inr, and the A nucleotide in the Inr consensus is usually designed as the “+ 1” position, whether or not transcription actually initiates at that particular nucleotide. This convention is useful because other core promoter motifs, such as the MTE and DPE, function with the Inr in a manner that exhibits a strict spacing dependence with the Inr consensus sequence (and hence, the A + 1 nucleotide) rather than the actual transcription start site (Burke and Kadonaga, 1997, Kutach and Kadonaga, 2000 and Lim et al., 2004)."[5]
"With TATA-driven core promoters, transcription can be achieved in vitro with purified RNA polymerase II, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH."[5]
"NC2 (negative cofactor 2; also known as Dr1-Drap1) [...] was identified as repressor of TATA-dependent transcription [...]."[5]
"TBP (TATA box-binding protein) activates TATA transcription [...] The TBP subunit binds to the TATA box [...] TFIIA appears to promote the binding of TBP to the TATA box."[5]
TATA boxes
"The TATA box is the first core promoter motif that was discovered (Goldberg, 1979) as well as the best known core promoter element. The metazoan TATA box consensus is TATAWAAR, where the upstream T is usually located at − 31 or − 30 relative to the A + 1 (or G + 1) position in the Inr (Carninci et al., 2006 and Ponjavic et al., 2006). [The] TATA box is recognized and bound by the TBP subunit of the TFIID complex. Both the TATA box and TBP are conserved from archaebacteria to humans (Reeve, 2003). The TATA box is also present in plants (Molina and Grotewold, 2005, Yamamoto et al., 2007a and Yamamoto et al., 2007b). Although the TATA box is a well known core promoter motif, it is present in only about 10%–15% of mammalian core promoters (Carninci et al., 2006, Kim et al., 2005 and Cooper et al., 2006)."[5]
"The BRE (TFIIBrecognition element) was initially identified as a TFIIB binding sequence that is immediately upstream of a subset (∼ 10%–30%) of TATA box elements (Lagrange et al., 1998). In addition, a second TFIIB recognition site, the BREd (downstream TFIIB recognition element), was found immediately downstream of the TATA box (Deng and Roberts, 2005). The discovery of the BREd led to the renaming of the original BRE as BREu for upstream BRE (reviewed in Deng and Roberts, 2007). Both the BREu and BREd function in conjunction with a TATA box and have been found to increase as well as to decrease the levels of basal transcription ( Lagrange et al., 1998, Evans et al., 2001 and Deng and Roberts, 2005). More recent studies suggest a distinct role for the BREu in transcriptional regulation (Juven-Gershon et al., 2008a; [...])."[5]
"TRF3 (also known as TBP2 and TBPL2) appears to be present only in vertebrates and is the TRF that is most closely related to TBP. TRF3 can bind to TATA boxes and support TATA-dependent transcription (Bártfai et al., 2004 and Jallow et al., 2004). TRF3 was found to be important for embryonic development (Bártfai et al., 2004 and Jallow et al., 2004). In addition, zebrafish embryos that are depleted of TRF3 exhibit multiple developmental defects and fail to undergo hematopoiesis (Hart et al., 2007)."[5]
"The top six sequences form a cohort of the eight-member consensus TATA(A/T)A(A/T)(A/G). Two missing members are TATA(A/T)ATG. This sequence ends in an ATG translational start codon, and thus is expected to be underrepresented in promoters. Since it is nevertheless part of the larger consensus that avidly binds TBP, this sequence was included in the TATA consensus, although it is rarely used."[8]
Hypotheses
- A1BG is not transcribed using a TATA box.
TATA box (Butler 2002) samplings
For the Basic programs testing consensus sequence TATAAA (starting with SuccessablesTATAB.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction: 2, TATAAA at 2852, TATAAA at 1602.
- Positive strand, negative direction: 3, TATAAA at 2874, TATAAA at 221, TATAAA at 182.
- negative strand, positive direction: 0.
- positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction: 3, TTTATA at 2869, TTTATA at 2638, TTTATA at 1740.
- inverse complement, positive strand, negative direction: 1, TTTATA at 219.
- inverse complement, negative strand, positive direction: 1, TTTATA at 2588.
- inverse complement, positive strand, positive direction: 0.
TATAB (4560-2846) UTRs
- Negative strand, negative direction: TATAAA at 2852.
- Negative strand, negative direction: TTTATA at 2869.
- Positive strand, negative direction: TATAAA at 2874.
TATAB negative direction (2811-2596) proximal promoters
- Negative strand, negative direction: TTTATA at 2638.
TATAB negative direction (2596-1) distal promoters
- Negative strand, negative direction: TATAAA at 1602.
- Negative strand, negative direction: TTTATA at 1740.
- Positive strand, negative direction: TATAAA at 221, TATAAA at 182.
- Positive strand, negative direction: TTTATA at 219.
TATAB positive direction (4050-1) distal promoters
- Negative strand, positive direction: TTTATA at 2588.
TATA box (Butler 2002) random dataset samplings
- TATABr0: 2, TATAAA at 3565, TATAAA at 499.
- TATABr1: 0.
- TATABr2: 1, TATAAA at 3856.
- TATABr3: 1, TATAAA at 4444.
- TATABr4: 2, TATAAA at 3685, TATAAA at 733.
- TATABr5: 1, TATAAA at 1563.
- TATABr6: 0.
- TATABr7: 1, TATAAA at 3629.
- TATABr8: 2, TATAAA at 706, TATAAA at 555.
- TATABr9: 3, TATAAA at 4219, TATAAA at 3150, TATAAA at 2621.
- TATABr0ci: 2, TTTATA at 1139, TTTATA at 497.
- TATABr1ci: 0.
- TATABr2ci: 0.
- TATABr3ci: 0.
- TATABr4ci: 1, TTTATA at 4527.
- TATABr5ci: 1, TTTATA at 2178.
- TATABr6ci: 0.
- TATABr7ci: 2, TTTATA at 4252, TTTATA at 2452.
- TATABr8ci: 1, TTTATA at 4222.
- TATABr9ci: 2, TTTATA at 3988, TTTATA at 162.
TATABr arbitrary (evens) (4560-2846) UTRs
- TATABr0: TATAAA at 3565.
- TATABr2: TATAAA at 3856.
- TATABr4: TATAAA at 3685.
- TATABr4ci: TTTATA at 4527.
- TATABr8ci: TTTATA at 4222.
TATABr alternate (odds) (4560-2846) UTRs
- TATABr3: TATAAA at 4444.
- TATABr9: TATAAA at 4219, TATAAA at 3150.
- TATABr7ci: TTTATA at 4252, TTTATA at 2452.
- TATABr9ci: TTTATA at 3988.
TATABr arbitrary positive direction (odds) (4445-4265) core promoters
- TATABr3: TATAAA at 4444.
TATABr arbitrary positive direction (odds) (4265-4050) proximal promoters
- TATABr9: TATAAA at 4219.
- TATABr7ci: TTTATA at 4252.
TATABr alternate positive direction (evens) (4265-4050) proximal promoters
- TATABr8ci: TTTATA at 4222.
TATABr arbitrary negative direction (evens) (2596-1) distal promoters
- TATABr0: TATAAA at 499.
- TATABr4: TATAAA at 733.
- TATABr8: TATAAA at 706, TATAAA at 555.
- TATABr0ci: TTTATA at 1139, TTTATA at 497.
TATABr alternate negative direction (odds) (2596-1) distal promoters
- TATABr5: TATAAA at 1563.
- TATABr5ci: TTTATA at 2178.
- TATABr7ci: TTTATA at 2452.
- TATABr9ci: TTTATA at 162.
TATABr arbitrary positive direction (odds) (4050-1) distal promoters
- TATABr5: TATAAA at 1563.
- TATABr9: TATAAA at 3150, TATAAA at 2621.
- TATABr5ci: TTTATA at 2178.
- TATABr7ci: TTTATA at 2452.
- TATABr9ci: TTTATA at 3988, TTTATA at 162.
TATABr alternate positive direction (evens) (4050-1) distal promoters
- TATABr0: TATAAA at 3565, TATAAA at 499.
- TATABr2: TATAAA at 3856.
- TATABr4: TATAAA at 3685, TATAAA at 733.
- TATABr8: TATAAA at 706, TATAAA at 555.
- TATABr0ci: TTTATA at 1139, TTTATA at 497.
TATA box (Butler 2002) analysis and results
TATA box consensus sequence TATAAA.[9]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 3 | 2 | 1.5 | 1.5 ± 0.5 (--2,+-1) |
| Randoms | UTR | arbitrary negative | 5 | 10 | 0.5 | 0.55 |
| Randoms | UTR | alternate negative | 6 | 10 | 0.6 | 0.55 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 1 | 10 | 0.1 | 0.05 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0.05 |
| Reals | Proximal | negative | 1 | 2 | 0.5 | 0.5 ± 0.5 (--1,+-0) |
| Randoms | Proximal | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 2 | 10 | 0.2 | 0.15 |
| Randoms | Proximal | alternate positive | 1 | 10 | 0.1 | 0.15 |
| Reals | Distal | negative | 5 | 2 | 2.5 | 2.5 ± 0.5 (--2,+-3) |
| Randoms | Distal | arbitrary negative | 6 | 10 | 0.6 | 0.5 |
| Randoms | Distal | alternate negative | 4 | 10 | 0.4 | 0.5 |
| Reals | Distal | positive | 1 | 2 | 0.5 | 0.5 ± 0.5 (-+1,++0) |
| Randoms | Distal | arbitrary positive | 7 | 10 | 0.7 | 0.8 |
| Randoms | Distal | alternate positive | 9 | 10 | 0.9 | 0.8 |
Comparison:
The occurrences of real TATAB UTRs, proximals and distals are greater than the randoms. This suggests that the real TATABs are likely active or activable.
Butler TATA box genes
- Gene ID: 2 v Z11711. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 70. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 133. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 183. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 185. It has a TATA box (TATAAA) from -6 to -1 nts from the TSS.[7]
- Gene ID: 203. It has a TATA box (TATAAA) from -4 to 2 nts from the TSS.[7]
- Gene ID: 265. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 279. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 280. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 292. It has a TATA box (TATAAA) from 3 to 8 nts from the TSS.[7]
- Gene ID: 314. It has a TATA box (TATAAA) from -41 to -36 nts from the TSS.[7]
- Gene ID: 338. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 358. It has a TATA box (TATAAA) from 23 to 28 nts from the TSS.[7]
- Gene ID: 358. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 359. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 360. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 374. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 383. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 390. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 468. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 482. It has a TATA box (TATAAA) from -35 to -30 nts from the TSS.[7]
- Gene ID: 496. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 677. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 694. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 846. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 1051. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 1101. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 1116. It has a TATA box (TATAAA) from 18 to 23 nts from the TSS.[7]
- Gene ID: 1152. It has a TATA box (TATAAA) from -41 to -36 nts from the TSS.[7]
- Gene ID: 1160. It has a TATA box (TATAAA) from -37 to -32 nts from the TSS.[7]
- Gene ID: 1180. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 1191. It has a TATA box (TATAAA) from -23 to -18 nts from the TSS.[7]
- Gene ID: 1231. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 1278. It has a TATA box (TATAAA) from -34 to -29 nts from the TSS.[7]
- Gene ID: 1401. It has a TATA box (TATAAA) from -45 to -40 nts from the TSS.[7]
- Gene ID: 1411. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 1427. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 1440. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 1543. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 1548. It has a TATA box (TATAAA) from -43 to -38 nts from the TSS.[7]
- Gene ID: 1553. It has a TATA box (TATAAA) from -45 to -40 nts from the TSS.[7]
- Gene ID: 1576. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 1735. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 1831. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 1833. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 1974. It has a TATA box (TATAAA) from 25 to 30 nts from the TSS.[7]
- Gene ID: 1990. It has a TATA box (TATAAA) from -23 to -18 nts from the TSS.[7]
- Gene ID: 1999. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 2206. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2250. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 2321. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 2542. It has a TATA box (TATAAA) from 28 to 33 nts from the TSS.[7]
- Gene ID: 2597. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2652. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2669. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2780. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2922. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 2947. It has a TATA box (TATAAA) from -43 to -38 nts from the TSS.[7]
- Gene ID: 3008. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 3015. It has a TATA box (TATAAA) from -8 to -3 nts from the TSS.[7]
- Gene ID: 3133. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 3158. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 3164. It has a TATA box (TATAAA) from -40 to -35 nts from the TSS.[7]
- Gene ID: 3171. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 3182. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 3206. It has a TATA box (TATAAA) from -39 to -34 nts from the TSS.[7]
- Gene ID: 3222. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 3232. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 3383. It has a TATA box (TATAAA) from -14 to -9 nts from the TSS.[7]
- Gene ID: 3458. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 3487. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 3558. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 3630. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 3640. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 3662. It has a TATA box (TATAAA) from -20 to -15 nts from the TSS.[7]
- Gene ID: 3726. It has a TATA box (TATAAA) from -42 to -37 nts from the TSS.[7]
- Gene ID: 3758. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 3858. It has a TATA box (TATAAA) from -34 to -29 nts from the TSS.[7]
- Gene ID: 3861. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 3872. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 3906. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 3938. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 3976. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 4049. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 4144. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 4222. It has a TATA box (TATAAA) from -21 to -16 nts from the TSS.[7]
- Gene ID: 4225. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 4284. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 4316. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 4321. It has a TATA box (TATAAA) from 5 to 10 nts from the TSS.[7]
- Gene ID: 4327. It has a TATA box (TATAAA) from -10 to -5 nts from the TSS.[7]
- Gene ID: 4357. It has a TATA box (TATAAA) from -17 to -12 nts from the TSS.[7]
- Gene ID: 4501. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 4582. It has a TATA box (TATAAA) from -24 to -19 nts from the TSS.[7]
- Gene ID: 4618. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 4624. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 4629. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 4638. It has a TATA box (TATAAA) from 30 to 35 nts from the TSS.[7]
- Gene ID: 4653. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 4741. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 4747. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 4843. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 4946. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 5004. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 5005. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 5016. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 5034. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 5052. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 5069. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 5132. It has a TATA box (TTTAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 5155. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 5224. It has a TATA box (TTTAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 5275. It has a TATA box (TATAAA) from -9 to -4 nts from the TSS.[7]
- Gene ID: 5360. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 5449. It has a TATA box (TATAAA) from 8 to 13 nts from the TSS.[7]
- Gene ID: 5478. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 5610. It has a TATA box (TATAAA) from 3 to 8 nts from the TSS.[7]
- Gene ID: 5617. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 5645. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 5650. It has a TATA box (TATAAA) from -2 to 4 nts from the TSS.[7]
- Gene ID: 5950. It has a TATA box (TATAAA) from -2 to 4 nts from the TSS.[7]
- Gene ID: 5956. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 5967. It has a TATA box (TATAAA) from 4 to 9 nts from the TSS.[7]
- Gene ID: 5996. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 6046. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 6280. It has a TATA box (TATAAA) from -23 to -18 nts from the TSS.[7]
- Gene ID: 6288. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 6289. It has a TATA box (TATAAA) from -17 to -12 nts from the TSS.[7]
- Gene ID: 6349. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 6351. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 6352. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 6364. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 6414. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 6427. It has a TATA box (TATAAA) from 11 to 16 nts from the TSS.[7]
- Gene ID: 6428. It has a TATA box (TATAAA) from 19 to 24 nts from the TSS.[7]
- Gene ID: 6435. It has a TATA box (TATAAA) from -42 to -37 nts from the TSS.[7]
- Gene ID: 6436. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 6446. It has a TATA box (TATAAA) from -17 to -12 nts from the TSS.[7]
- Gene ID: 6500. It has a TATA box (TATAAA) from 1 to 6 nts from the TSS.[7]
- Gene ID: 6513. It has a TATA box (TATAAA) from 18 to 23 nts from the TSS.[7]
- Gene ID: 6519. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 6647. It has a TATA box (TATAAA) from -34 to -29 nts from the TSS.[7]
- Gene ID: 6707. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 6783. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 6870. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 6916. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 7037. It has a TATA box (TATAAA) from 4 to 9 nts from the TSS.[7]
- Gene ID: 7042. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 7056. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 7124. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 7252. It has a TATA box (TATAAA) from -35 to -30 nts from the TSS.[7]
- Gene ID: 7262. It has a TATA box (TATAAA) from -31 to -16 nts from the TSS.[7]
- Gene ID: 7295. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 7295. It has a TATA box (TATAAA) from 25 to 30 nts from the TSS.[7]
- Gene ID: 7432. It has a TATA box (TATAAA) from -45 to -40 nts from the TSS.[7]
- Gene ID: 7803. It has a TATA box (TATAAA) from -25 to -20 nts from the TSS.[7]
- Gene ID: 7850. It has a TATA box (TATAAA) from 23 to -28 nts from the TSS.[7]
- Gene ID: 8000. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 8431. It has a TATA box (TATAAA) from -35 to -30 nts from the TSS.[7]
- Gene ID: 8942. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 8991. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 4327. It has a TATA box (TATAAA) from -10 to -5 nts from the TSS.[7]
- Gene ID: 9001. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 9518. It has a TATA box (TATAAA) from -24 to -19 nts from the TSS.[7]
- Gene ID: 9643. It has a TATA box (TATAAA) from 18 to 23 nts from the TSS.[7]
- Gene ID: 9768. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 10057. It has a TATA box (TATAAA) from 8 to 13 nts from the TSS.[7]
- Gene ID: 10284. It has a TATA box (TATAAA) from -22 to -17 nts from the TSS.[7]
- Gene ID: 10397. It has a TATA box (TATAAA) from -21 to -16 nts from the TSS.[7]
- Gene ID: 10458. It has a TATA box (TATAAA) from -34 to -29 nts from the TSS.[7]
- Gene ID: 10482. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 10563. It has a TATA box (TATAAA) from 33 to 38 nts from the TSS.[7]
- Gene ID: 10631. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 10761. It has a TATA box (TATAAA) from -43 to -38 nts from the TSS.[7]
- Gene ID: 10761. It has a TATA box (TATAAA) from -2 to 4 nts from the TSS.[7]
- Gene ID: 10930. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 10938. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 11081. It has a TATA box (TATAAA) from -38 to -33 nts from the TSS.[7]
- Gene ID: 22928. It has a TATA box (TATAAA) from -33 to -28 nts from the TSS.[7]
- Gene ID: 22943. It has a TATA box (TATAAA) from 42 to 47 nts from the TSS.[7]
- Gene ID: 25928. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 26287. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 26576. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 26827. There is a Hogness box sequence TATAAA beginning at position -31.
- Gene ID: 27129. It has a TATA box (TATAAA) from -15 to -10 nts from the TSS.[7]
- Gene ID: 27316. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 51155. It has a TATA box (TATAAA) from -20 to -15 nts from the TSS.[7]
- Gene ID: 51203. It has a TATA box (TATAAA) from -20 to -15 nts from the TSS.[7]
- Gene ID: 51297. It has a TATA box (TATAAA) from -45 to -40 nts from the TSS.[7]
- Gene ID: 51313. It has a TATA box (TATAAA) from 2 to 7 nts from the TSS.[7]
- Gene ID: 51582. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 55118. It has a TATA box (TATAAA) from 1 to 6 nts from the TSS.[7]
- Gene ID: 55504. It has a TATA box (TATAAA) from -26 to -21 nts from the TSS.[7]
- Gene ID: 56642. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 56895. It has a TATA box (TATAAA) from 40 to 45 nts from the TSS.[7]
- Gene ID: 57126. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 57152. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 57579. It has a TATA box (TATAAA) from -16 to -11 nts from the TSS.[7]
- Gene ID: 80740. It has a TATA box (TATAAA) from -44 to -39 nts from the TSS.[7]
- Gene ID: 81606. It has a TATA box (TATAAA) from -29 to -24 nts from the TSS.[7]
- Gene ID: 83638. It has a TATA box (TATAAA) from -6 to -1 nts from the TSS.[7]
- Gene ID: 84223. It has a TATA box (TATAAA) from 26 to 31 nts from the TSS.[7]
- Gene ID: 92736. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 115265. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
- Gene ID: 117156. It has a TATA box (TATAAA) from -31 to -26 nts from the TSS.[7]
- Gene ID: 126364. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 145957. It has a TATA box (TATAAA) from -30 to -25 nts from the TSS.[7]
- Gene ID: 153020. It has a TATA box (TATAAA) from 8 to 13 nts from the TSS.[7]
- Gene ID: 200504. It has a TATA box (TATAAA) from -27 to -22 nts from the TSS.[7]
- Gene ID: 200539. It has a TATA box (TATAAA) from -32 to -27 nts from the TSS.[7]
- Gene ID: 342574. It has a TATA box (TATAAA) from -28 to -23 nts from the TSS.[7]
TATA boxes (Carninci 2006) samplings
For the Basic programs testing consensus sequence TATAAAA (starting with SuccessablesTATACA--.bas.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- negative strand, negative direction: 1, TATAAAA at 2853.
- positive strand, negative direction: 2, TATAAAA at 222, TATAAAA at 183.
- negative strand, positive direction: 0.
- positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction: 2, TTTTATA at 2869, TTTTATA at 1740.
- inverse complement, positive strand, negative direction: 1, TTTTATA at 219.
- inverse complement, negative strand, positive direction: 0.
- inverse complement, positive strand, positive direction: 0.
TATAAAA (4560-2846) UTRs
- Negative strand, negative direction: TATAAAA at 2853.
- Negative strand, negative direction: TTTTATA at 2869.
TATAAAA negative direction (2596-1) distal promoters
- Negative strand, negative direction: TTTTATA at 1740.
- Positive strand, negative direction: TATAAAA at 222, TATAAAA at 183.
- Positive strand, negative direction: TTTTATA at 219.
TATA boxes (Carninci 2006) random dataset samplings
- TATACr0: 0.
- TATACr1: 0.
- TATACr2: 0.
- TATACr3: 1, TATAAAA at 4445.
- TATACr4: 0.
- TATACr5: 0.
- TATACr6: 0.
- TATACr7: 0.
- TATACr8: 1, TATAAAA at 707.
- TATACr9: 1, TATAAAA at 2622.
- TATACr0ci: 1, TTTTATA at 497.
- TATACr1ci: 0.
- TATACr2ci: 0.
- TATACr3ci: 0.
- TATACr4ci: 1, TTTTATA at 4527.
- TATACr5ci: 0.
- TATACr6ci: 0.
- TATACr7ci: 0.
- TATACr8ci: 0.
- TATACr9ci: 1, TTTTATA at 162.
TATACr arbitrary (evens) (4560-2846) UTRs
- TATACr4ci: TTTTATA at 4527.
TATACr alternate (odds) (4560-2846) UTRs
- TATACr3: TATAAAA at 4445.
TATACr arbitrary positive direction (odds) (4445-4265) core promoters
- TATACr3: TATAAAA at 4445.
TATACr alternate negative direction (odds) (2811-2596) proximal promoters
- TATACr9: TATAAAA at 2622.
TATACr arbitrary negative direction (evens) (2596-1) distal promoters
- TATACr8: TATAAAA at 707.
- TATACr0ci: TTTTATA at 497.
TATACr alternate negative direction (odds) (2596-1) distal promoters
- TATACr9ci: TTTTATA at 162.
TATACr arbitrary positive direction (odds) (4050-1) distal promoters
- TATACr9: TATAAAA at 2622.
- TATACr9ci: TTTTATA at 162.
TATACr alternate positive direction (evens) (4050-1) distal promoters
- TATACr8: TATAAAA at 707.
- TATACr0ci: TTTTATA at 497.
TATAAAA (Carninci 2006) analysis and results
A genome-wide study put the fraction of TATAAAA-dependent human promoters at ~10%.[10]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 2 | 2 | 1 | 1 ± 1 (--2,+-0) |
| Randoms | UTR | arbitrary negative | 1 | 10 | 0.1 | 0.1 |
| Randoms | UTR | alternate negative | 1 | 10 | 0.1 | 0.1 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 1 | 10 | 0.1 | 0.05 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0.05 |
| Reals | Proximal | negative | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary negative | 0 | 10 | 0 | 0.05 |
| Randoms | Proximal | alternate negative | 1 | 10 | 0.1 | 0.05 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate positive | 0 | 10 | 0 | 0 |
| Reals | Distal | negative | 4 | 2 | 2 | 2 ± 1 (--1,+-3) |
| Randoms | Distal | arbitrary negative | 2 | 10 | 0.2 | 0.15 |
| Randoms | Distal | alternate negative | 1 | 10 | 0.1 | 0.15 |
| Reals | Distal | positive | 0 | 2 | 0 | 0 |
| Randoms | Distal | arbitrary positive | 2 | 10 | 0.2 | 0.2 |
| Randoms | Distal | alternate positive | 2 | 10 | 0.2 | 0.2 |
Comparison:
The occurrences of real TATACs are greater than the randoms. This suggests that the real TATACs are likely active or activable.
Carninci TATA box genes
- Gene ID: 19. It has a TATA box (TATAAAA) from -34 to -28 nts from the TSS.[7]
- Gene ID: 58. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 60. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 173. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 174. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 229. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 229. It has a TATA box (TATAAAA) from 23 to 29 nts from the TSS.[7]
- Gene ID: 230. It has a TATA box (TATAAAA) from -19 to -13 nts from the TSS.[7]
- Gene ID: 301. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 302. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 345. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 467. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 676. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 759. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 760. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 762. It has a TATA box (TATAAAA) from 2 to 8 nts from the TSS.[7]
- Gene ID: 767. It has a TATA box (TATAAAA) from -34 to -28 nts from the TSS.[7]
- Gene ID: 811. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 1044. It has a TATA box (TATAAAA) from -34 to -28 nts from the TSS.[7]
- Gene ID: 1081. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 1215. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 1277. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 1356. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 1356. It has a TATA box (TATAAAA) from 27 to 33 nts from the TSS.[7]
- Gene ID: 1382. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 1392. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 1393. It has a TATA box (TATAAAA) from 36 to 42 nts from the TSS.[7]
- Gene ID: 1393. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 1490. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 1544. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 1571. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 1581. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 1655. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 1805. It has a TATA box (TATAAAA) from -26 to -20 nts from the TSS.[7]
- Gene ID: 1893. It has a TATA box (TATAAAA) from 17 to 23 nts from the TSS.[7]
- Gene ID: 1906. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 1938. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 1974. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 2168. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 2353. It has a TATA box (TATAAAA) from -22 to -16 nts from the TSS.[7]
- Gene ID: 2538. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 2641. It has a TATA box (TATAAAA) from -25 to -19 nts from the TSS.[7]
- Gene ID: 2688. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 3273. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 3304. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 3397. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 3491. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 3596. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 3605. It has a TATA box (TATAAAA) from -44 to -38 nts from the TSS.[7]
- Gene ID: 3624. It has a TATA box (TATAAAA) from 7 to 13 nts from the TSS.[7]
- Gene ID: 3860. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 4025. It has a TATA box (TATAAAA) from 11 to 17 nts from the TSS.[7]
- Gene ID: 4254. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 4256. It has a TATA box (TATAAAA) from 2 to 8 nts from the TSS.[7]
- Gene ID: 4319. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 4322. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 4435. It has a TATA box (TATAAAA) from -2 to 5 nts from the TSS.[7]
- Gene ID: 4504. It has a TATA box (TATAAAA) from -24 to -18 nts from the TSS.[7]
- Gene ID: 4609. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 4616. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 4744. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 4842. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 4856. It has a TATA box (TATAAAA) from -291 to -23 nts from the TSS.[7]
- Gene ID: 4878. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 5033. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 5054. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 5055. It has a TATA box (TATAAAA) from -26 to -20 nts from the TSS.[7]
- Gene ID: 5068. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 5079. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 5225. It has a TATA box (TATAAAA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 5406. It has a TATA box (TATAAAA) from -24 to -18 nts from the TSS.[7]
- Gene ID: 5408. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 5478. It has a TATA box (TATAAAA) from 8 to 14 nts from the TSS.[7]
- Gene ID: 5514. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 5741. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 5743. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 6175. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 6187. It has a TATA box (TATAAAA) from -25 to -19 nts from the TSS.[7]
- Gene ID: 6279. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 6354. It has a TATA box (TATAAAA) from -17 to -11 nts from the TSS.[7]
- Gene ID: 6356. It has a TATA box (TATAAAA) from -44 to -38 nts from the TSS.[7]
- Gene ID: 6357. It has a TATA box (TATAAAA) from -36 to -30 nts from the TSS.[7]
- Gene ID: 6376. It has a TATA box (TATAAAA) from -12 to -6 nts from the TSS.[7]
- Gene ID: 6518. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 6559. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 6624. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 6698. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 6781. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 6906. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 6988. It has a TATA box (TATAAAA) from -26 to -20 nts from the TSS.[7]
- Gene ID: 7031. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 7032. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 7038. It has a TATA box (TATAAAA) from -35 to -29 nts from the TSS.[7]
- Gene ID: 7043. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 7252. It has a TATA box (TATAAAA) from 2 to 8 nts from the TSS.[7]
- Gene ID: 7276. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 7280. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 7306. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 7432. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 7803. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 7852. It has a TATA box (TATAAAA) from -26 to -20 nts from the TSS.[7]
- Gene ID: 8288. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 8339. It has a TATA box (TATAAAA) from -33 to -27 nts from the TSS.[7]
- Gene ID: 8564. It has a TATA box (TATAAAA) from -14 to -8 nts from the TSS.[7]
- Gene ID: 8832. It has a TATA box (TATAAAA) from -12 to -6 nts from the TSS.[7]
- Gene ID: 8969. It has a TATA box (TATAAAA) from -27 to -21 nts from the TSS.[7]
- Gene ID: 8970. It has a TATA box (TATAAAA) from -28 to -22 nts from the TSS.[7]
- Gene ID: 9510. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 9607. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 9709. It has a TATA box (TATAAAA) from 1 to 7 nts from the TSS.[7]
- Gene ID: 10215. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 10350. It has a TATA box (TATAAAA) from 31 to 37 nts from the TSS.[7]
- Gene ID: 11067. It has a TATA box (TATAAAA) from 14 to 20 nts from the TSS.[7]
- Gene ID: 11082. It has a TATA box (TATAAAA) from -11 to -5 nts from the TSS.[7]
- Gene ID: 11169. It has a TATA box (TATAAAA) from -16 to -10 nts from the TSS.[7]
- Gene ID: 23645. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 27106. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 50943. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 51050. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 51129. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 51297. It has a TATA box (TATAAAA) from -45 to -39 nts from the TSS.[7]
- Gene ID: 54106. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 55603. It has a TATA box (TATAAAA) from -2 to 5 nts from the TSS.[7]
- Gene ID: 56675. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 56829. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 56987. It has a TATA box (TATAAAA) from -35 to -29 nts from the TSS.[7]
- Gene ID: 57626. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 57823. It has a TATA box (TATAAAA) from -15 to -9 nts from the TSS.[7]
- Gene ID: 64111. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 80177. It has a TATA box (TATAAAA) from -34 to -28 nts from the TSS.[7]
- Gene ID: 81285. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 83998. It has a TATA box (TATAAAA) from -29 to -23 nts from the TSS.[7]
- Gene ID: 84107. It has a TATA box (TATAAAA) from 29 to 35 nts from the TSS.[7]
- Gene ID: 85235. It has a TATA box (TATAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 114899. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 116842. It has a TATA box (TATAAAA) from -32 to -26 nts from the TSS.[7]
- Gene ID: 130120. It has a TATA box (TATAAAA) from 17 to 23 nts from the TSS.[7]
- Gene ID: 145957. It has a TATA box (TATAAAA) from -7 to -1 nts from the TSS.[7]
- Gene ID: 147183. It has a TATA box (TATAAAA) from -30 to -24 nts from the TSS.[7]
TATA box (Watson 2014) samplings
For the Basic programs testing consensus sequence TATA(A/T)A(A/T) (starting with SuccessablesTATAW.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction: 4, TATATAT at 2872, TATAAAA at 2853, TATATAA at 1601, TATATAT at 1599.
- Positive strand, negative direction: 3, TATATAA at 2873, TATATAT at 1600, TATAAAA at 222, TATAAAA at 183.
- Negative strand, positive direction: 0.
- Positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction: 5, TTATATA at 2871, TTTTATA at 2869, ATTTATA at 2638, TTTTATA at 1740.
- inverse complement, positive strand, negative direction: 2, ATATATA at 1599, TTTTATA at 219.
- inverse complement, negative strand, positive direction: 0.
- inverse complement, positive strand, positive direction: 0.
TATAW (4560-2846) UTRs
- Negative strand, negative direction: TATATAT at 2872, TATAAAA at 2853.
- Negative strand, negative direction: TTATATA at 2871, TTTTATA at 2869.
- Positive strand, negative direction: TATATAA at 2873.
TATAW negative direction (2811-2596) proximal promoters
- Negative strand, negative direction: ATTTATA at 2638.
TATAW negative direction (2596-1) distal promoters
- Negative strand, negative direction: TATATAA at 1601, TATATAT at 1599.
- Negative strand, negative direction: TTTTATA at 1740.
- Positive strand, negative direction: TATATAT at 1600, TATAAAA at 222, TATAAAA at 183.
- Positive strand, negative direction: TTTTATA at 219.
TATA box (Watson 2014) random dataset samplings
- TATAWr0: 1, TATAAAT at 3566.
- TATAWr1: 1, TATATAT at 139.
- TATAWr2: 0.
- TATAWr3: 1, TATAAAA at 4445.
- TATAWr4: 0.
- TATAWr5: 1, TATAAAT at 1564.
- TATAWr6: 0.
- TATAWr7: 1, TATAAAT at 3630.
- TATAWr8: 1, TATAAAA at 707.
- TATAWr9: 2, TATATAA at 3149, TATAAAA at 2622.
- TATAWr0ci: 2, ATATATA at 2701, TTTTATA at 497.
- TATAWr1ci: 1, TTATATA at 1267.
- TATAWr2ci: 0.
- TATAWr3ci: 1, ATATATA at 3803.
- TATAWr4ci: 2, TTTTATA at 4527, TTATATA at 4254.
- TATAWr5ci: 0.
- TATAWr6ci: 1, TTATATA at 833.
- TATAWr7ci: 1, TTATATA at 4254.
- TATAWr8ci: 0.
- TATAWr9ci: 1, TTTTATA at 162.
TATAWr arbitrary (evens) (4560-2846) UTRs
- TATAWr0: TATAAAT at 3566.
- TATAWr4ci: TTTTATA at 4527, TTATATA at 4254.
TATAWr alternate (odds) (4560-2846) UTRs
- TATAWr3: TATAAAA at 4445.
- TATAWr7: TATAAAT at 3630.
- TATAWr9: TATATAA at 3149.
- TATAWr3ci: ATATATA at 3803.
- TATAWr7ci: TTATATA at 4254.
TATAWr arbitrary positive direction (odds) (4445-4265) core promoters
- TATAWr3: TATAAAA at 4445.
TATAWr arbitrary negative direction (evens) (2811-2596) proximal promoters
- TATAWr0ci: ATATATA at 2701.
TATAWr alternate negative direction (odds) (2811-2596) proximal promoters
- TATAWr9: TATAAAA at 2622.
TATAWr arbitrary positive direction (odds) (4265-4050) proximal promoters
- TATAWr7ci: TTATATA at 4254.
TATAWr alternate positive direction (evens) (4265-4050) proximal promoters
- TATAWr4ci: TTATATA at 4254.
TATAWr arbitrary negative direction (evens) (2596-1) distal promoters
- TATAWr8: TATAAAA at 707.
- TATAWr0ci: TTTTATA at 497.
- TATAWr6ci: TTATATA at 833.
TATAWr alternate negative direction (odds) (2596-1) distal promoters
- TATAWr1: TATATAT at 139.
- TATAWr5: TATAAAT at 1564.
- TATAWr1ci: TTATATA at 1267.
- TATAWr9ci: TTTTATA at 162.
TATAWr arbitrary positive direction (odds) (4050-1) distal promoters
- TATAWr1: TATATAT at 139.
- TATAWr5: TATAAAT at 1564.
- TATAWr7: TATAAAT at 3630.
- TATAWr9: TATATAA at 3149, TATAAAA at 2622.
- TATAWr1ci: TTATATA at 1267.
- TATAWr3ci: ATATATA at 3803.
- TATAWr9ci: TTTTATA at 162.
TATAWr alternate positive direction (evens) (4050-1) distal promoters
- TATAWr0: TATAAAT at 3566.
- TATAWr8: TATAAAA at 707.
- TATAWr0ci: ATATATA at 2701, TTTTATA at 497.
- TATAWr6ci: TTATATA at 833.
TATA box (Watson 2014) analysis and results
The TATA box is a component of the eukaryotic core promoter and generally contains the consensus sequence TATA(A/T)A(A/T).[11]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 5 | 2 | 2.5 | 2.5 ± 1.5 (--4,+-1) |
| Randoms | UTR | arbitrary negative | 3 | 10 | 0.3 | 0.4 |
| Randoms | UTR | alternate negative | 5 | 10 | 0.5 | 0.4 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 1 | 10 | 0.1 | 0.05 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0.05 |
| Reals | Proximal | negative | 1 | 2 | 0.5 | 0.5 |
| Randoms | Proximal | arbitrary negative | 1 | 10 | 0.1 | 0.1 |
| Randoms | Proximal | alternate negative | 1 | 10 | 0.1 | 0.1 |
| Reals | Proximal | positive | 1 | 2 | 0.5 | 0.5 ± 0.5 (-+1,++0) |
| Randoms | Proximal | arbitrary positive | 1 | 10 | 0.1 | 0.1 |
| Randoms | Proximal | alternate positive | 1 | 10 | 0.1 | 0.1 |
| Reals | Distal | negative | 7 | 2 | 3.5 | 3.5 ± 0.5 (--3,+-4) |
| Randoms | Distal | arbitrary negative | 3 | 10 | 0.3 | 0.35 |
| Randoms | Distal | alternate negative | 4 | 10 | 0.4 | 0.35 |
| Reals | Distal | positive | 0 | 2 | 0 | 0 |
| Randoms | Distal | arbitrary positive | 8 | 10 | 0.8 | 0.65 |
| Randoms | Distal | alternate positive | 5 | 10 | 0.5 | 0.65 |
Comparison:
The occurrences of real TATAW UTRs, proximals and distals are greater than the randoms. This suggests that the real TATAWs are likely active or activable.
Watson TATA box genes
- Gene ID: 262. It has a TATA box (TATATAAG) from 10 to 17 nts from the TSS.[7]
- Gene ID: 292. It has a TATA box (TATATAAA) from 1 to 8 nts from the TSS.[7]
- Gene ID: 604. It has a TATA box (TATATATA) from -22 to -15 nts from the TSS.[7]
- Gene ID: 794. It has a TATA box (TATATAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 1116. It has a TATA box (TATATAAA) from 16 to 23 nts from the TSS.[7]
- Gene ID: 1153. It has a TATA box (TATATAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 1158. It has a TATA box (TATATAAG) from -12 to -5 nts from the TSS.[7]
- Gene ID: 1410. It has a TATA box (TATATAAG) from -26 to -19 nts from the TSS.[7]
- Gene ID: 1581. It has a TATA box (TATATAAA) from -33 to -26 nts from the TSS.[7]
- Gene ID: 1728. It has a TATA box (TATATAAG) from -12 to -5 nts from the TSS.[7]
- Gene ID: 1805. It has a TATA box (TATATAAA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 1811. It has a TATA box (TATATATA) from -19 to -12 nts from the TSS.[7]
- Gene ID: 1831. It has a TATA box (TATATAAA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 1915. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 1961. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 2244. It has a TATA box (TATATATA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 2641. It has a TATA box (TATATAAA) from -27 to -20 nts from the TSS.[7]
- Gene ID: 2814. It has a TATA box (TATATATG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 3006. It has a TATA box (TATATATA) from -37 to -28 nts from the TSS.[7]
- Gene ID: 3008. It has a TATA box (TATATAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 3009. It has a TATA box (TATATAAG) from -38 to -31 nts from the TSS.[7]
- Gene ID: 3009. It has a TATA box (TATATAAG) from 29 to 36 nts from the TSS.[7]
- Gene ID: 3010. It has a TATA box (TATATAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 3050. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 3164. It has a TATA box (TATATAAA) from -42 to -35 nts from the TSS.[7]
- Gene ID: 3232. It has a TATA box (TATATAAA) from -33 to -26 nts from the TSS.[7]
- Gene ID: 3280. It has a TATA box (TATATATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 3309. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 3320. It has a TATA box (TATATAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 3371. It has a TATA box (TATATAAG) from -34 to -27 nts from the TSS.[7]
- Gene ID: 3375. It has a TATA box (TATATAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 3565. It has a TATA box (TATATATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 3593. It has a TATA box (TATATATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 3708. It has a TATA box (TATATAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 3848. It has a TATA box (TATATAAG) from -43 to -36 nts from the TSS.[7]
- Gene ID: 3858. It has a TATA box (TATATAAA) from -36 to -29 nts from the TSS.[7]
- Gene ID: 3859. It has a TATA box (TATATAAG) from -45 to -38 nts from the TSS.[7]
- Gene ID: 3976. It has a TATA box (TATATAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 3977. It has a TATA box (TATATATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 4014. It has a TATA box (TATATATA) from -40 to -33 nts from the TSS.[7]
- Gene ID: 4316. It has a TATA box (TATATAAA) from -27 to -20 nts from the TSS.[7]
- Gene ID: 4321. It has a TATA box (TATATAAA) from 3 to 10 nts from the TSS.[7]
- Gene ID: 4618. It has a TATA box (TATATATA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4632. It has a TATA box (TATATATG) from -27 to -20 nts from the TSS.[7]
- Gene ID: 4638. It has a TATA box (TATATATG) from 28 to 35 nts from the TSS.[7]
- Gene ID: 4653. It has a TATA box (TATATATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 4842. It has a TATA box (TATATAAA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 4869. It has a TATA box (TATATATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 4922. It has a TATA box (TATATATA) from -33 to -26 nts from the TSS.[7]
- Gene ID: 4982. It has a TATA box (TATATATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 5443. It has a TATA box (TATATAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 5449. It has a TATA box (TATATATG) from 18 to 25 nts from the TSS.[7]
- Gene ID: 5553. It has a TATA box (TATATAAG) from -34 to -27 nts from the TSS.[7]
- Gene ID: 5741. It has a TATA box (TATATATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 5744. It has a TATA box (TATATATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 5996. It has a TATA box (TATATAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 6046. It has a TATA box (TATATATA) from -37 to -30 nts from the TSS.[7]
- Gene ID: 6224. It has a TATA box (TATATAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 6232. It has a TATA box (TATATAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 6432. It has a TATA box (TATATAAA) from -9 to -2 nts from the TSS.[7]
- Gene ID: 6707. It has a TATA box (TATATAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 7021. It has a TATA box (TATATATG) from -48 to -41 nts from the TSS.[7]
- Gene ID: 7167. It has a TATA box (TATATAAG) from -2 to 5 nts from the TSS.[7]
- Gene ID: 7316. It has a TATA box (TATATAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 7369. It has a TATA box (TATATATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 7432. It has a TATA box (TATATAAA) from -47 to -40 nts from the TSS.[7]
- Gene ID: 8483. It has a TATA box (TATATAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 8490. It has a TATA box (TATATATA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 8513. It has a TATA box (TATATAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 8970. It has a TATA box (TATATAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 9421. It has a TATA box (TATATAAG) from 7 to 14 nts from the TSS.[7]
- Gene ID: 9921. It has a TATA box (TATATATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 10631. It has a TATA box (TATATAAA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 10761. It has a TATA box (TATATAAA) from -45 to -38 nts from the TSS.[7]
- Gene ID: 10769. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 10912. It has a TATA box (TATATAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 11009. It has a TATA box (TATATATG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 23450. It has a TATA box (TATATATA) from -40 to -33 nts from the TSS.[7]
- Gene ID: 25928. It has a TATA box (TATATAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 27063. It has a TATA box (TATATAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 51278. It has a TATA box (TATATAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 51738. It has a TATA box (TATATAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 54567. It has a TATA box (TATATATA) from -35 to -28 nts from the TSS.[7]
- Gene ID: 55504. It has a TATA box (TATATATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 56987. It has a TATA box (TATATAAA) from -37 to -29 nts from the TSS.[7]
- Gene ID: 57626. It has a TATA box (TATATAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 65108. It has a TATA box (TATATATA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 79733. It has a TATA box (TATATATA) from -12 to -5 nts from the TSS.[7]
- Gene ID: 84328. It has a TATA box (TATATATA) from -45 to -38 nts from the TSS.[7]
- Gene ID: 84419. It has a TATA box (TATATATA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 84790. It has a TATA box (TATATAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 84889. It has a TATA box (TATATAAG) from -40 to -33 nts from the TSS.[7]
- Gene ID: 131377. It has a TATA box (TATATATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 342574. It has a TATA box (TATATAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 389125. It has a TATA box (TATATAAG) from 13 to 20 nts from the TSS.[7]
TATA box (Juven-Gershon 2010) samplings
For the Basic programs (starting with SuccessablesTATAJ.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction, looking for TATA(A/T)AA(A/G): 1, TATATAAA at 1602.
- Positive strand, negative direction: 3, TATATAAA at 2874, TATAAAAG at 223, TATAAAAG at 184.
- Negative strand, positive direction: 0.
- Positive strand, positive direction: 0.
- complement, negative strand, negative direction: 3, ATATATTT at 2874, ATATTTTC at 223, ATATTTTC at 184.
- complement, positive strand, negative direction: 1, ATATATTT at 1602.
- complement, negative strand, positive direction: 0.
- complement, positive strand, positive direction: 0.
- inverse, negative strand, negative direction: AAAAATAT at 219
- inverse, positive strand, negative direction: 0
- inverse, negative strand, positive direction: 0.
- inverse, positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction, looking for (C/T)TT(A/T)TATA: 1, TTTATATA at 2871.
- inverse complement, positive strand, negative direction: 1, TTTTTATA at 219.
- inverse complement, negative strand, positive direction: 0.
- inverse complement, positive strand, positive direction: 0.
TATAJ (4560-2846) UTRs
- Negative strand, negative direction: TTTATATA at 2871.
- Positive strand, negative direction: TATATAAA at 2874.
TATAJ negative direction (2596-1) distal promoters
- Negative strand, negative direction: TATATAAA at 1602.
- Positive strand, negative direction: TATAAAAG at 223, TATAAAAG at 184.
- Positive strand, negative direction: TTTTTATA at 219.
TATA box (Juven-Gershon 2010) random dataset samplings
- TATAJr0: 0.
- TATAJr1: 0.
- TATAJr2: 0.
- TATAJr3: 0.
- TATAJr4: 0.
- TATAJr5: 0.
- TATAJr6: 0.
- TATAJr7: 0.
- TATAJr8: 1, TATAAAAA at 708.
- TATAJr9: 1, TATATAAA at 3150.
- TATAJr0ci: 0.
- TATAJr1ci: 0.
- TATAJr2ci: 0.
- TATAJr3ci: 0.
- TATAJr4ci: 2, CTTTTATA at 4527, CTTATATA at 4254.
- TATAJr5ci: 0.
- TATAJr6ci: 1, CTTATATA at 833.
- TATAJr7ci: 1, TTTATATA at 4254.
- TATAJr8ci: 0.
- TATAJr9ci: 0.
TATAJr arbitrary (evens) (4560-2846) UTRs
- TATAJr4ci: CTTTTATA at 4527, CTTATATA at 4254.
TATAJr alternate (odds) (4560-2846) UTRs
- TATAJr9: TATATAAA at 3150.
- TATAJr7ci: TTTATATA at 4254.
TATAJr alternate positive direction (evens) (4445-4265) core promoters
- TATAJr4ci: CTTATATA at 4254.
TATAJr arbitrary positive direction (odds) (4265-4050) proximal promoters
- TATAJr7ci: TTTATATA at 4254.
TATAJr alternate positive direction (evens) (4265-4050) proximal promoters
- TATAJr4ci: CTTATATA at 4254.
TATAJr arbitrary negative direction (evens) (2596-1) distal promoters
- TATAJr8: TATAAAAA at 708.
- TATAJr6ci: CTTATATA at 833.
TATAJr arbitrary positive direction (odds) (4050-1) distal promoters
- TATAJr9: TATATAAA at 3150.
TATAJr alternate positive direction (evens) (4050-1) distal promoters
- TATAJr8: TATAAAAA at 708.
- TATAJr6ci: CTTATATA at 833.
TATA box (Juven-Gershon 2010) analysis and results
"The metazoan TATA box consensus is TATAWAAR [...]."[5]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 2 | 2 | 1 | 1 ± 0 (--1,+-1) |
| Randoms | UTR | arbitrary negative | 2 | 10 | 0.2 | 0.2 |
| Randoms | UTR | alternate negative | 2 | 10 | 0.2 | 0.2 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 0 | 10 | 0 | 0.05 |
| Randoms | Core | alternate positive | 1 | 10 | 0.1 | 0.05 |
| Reals | Proximal | negative | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 1 | 10 | 0.1 | 0.1 |
| Randoms | Proximal | alternate positive | 1 | 10 | 0.1 | 0.1 |
| Reals | Distal | negative | 4 | 2 | 2 | 2 ± 1 (--1,+-3) |
| Randoms | Distal | arbitrary negative | 2 | 10 | 0.2 | 0.1 |
| Randoms | Distal | alternate negative | 0 | 10 | 0 | 0.1 |
| Reals | Distal | positive | 0 | 2 | 0 | 0 |
| Randoms | Distal | arbitrary positive | 1 | 10 | 0.1 | 0.15 |
| Randoms | Distal | alternate positive | 2 | 10 | 0.2 | 0.15 |
Comparison:
The occurrences of real TATAJ UTRs and distals are greater than the randoms. This suggests that the real TATAJs are likely active or activable.
Juven-Gershon TATA box genes
- Gene ID: 19. It has a TATA box (TATAAAAG) from -34 to -27 nts from the TSS.[7]
- Gene ID: 174. It has a TATA box (TATAAAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 302. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 467. It has a TATA box (TATAAAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 676. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 760. It has a TATA box (TATAAAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 811. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 1044. It has a TATA box (TATAAAAG) from -34 to -27 nts from the TSS.[7]
- Gene ID: 1081. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 1277. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 1382. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 1393. It has a TATA box (TATAAAAG) from 36 to 43 nts from the TSS.[7]
- Gene ID: 1393. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 1490. It has a TATA box (TATAAAAG) from -27 to -20 nts from the TSS.[7]
- Gene ID: 1655. It has a TATA box (TATAAAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 1893. It has a TATA box (TATAAAAG) from 17 to 24 nts from the TSS.[7]
- Gene ID: 2353. It has a TATA box (TATAAAAG) from -22 to -15 nts from the TSS.[7]
- Gene ID: 2641. It has a TATA box (TATAAAAG) from -25 to -18 nts from the TSS.[7]
- Gene ID: 3304. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 3397. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 3491. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 3596. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 3860. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 4025. It has a TATA box (TATAAAAG) from 11 to 18 nts from the TSS.[7]
- Gene ID: 4254. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4319. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4322. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4504. It has a TATA box (TATAAAAG) from -24 to -17 nts from the TSS.[7]
- Gene ID: 4609. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 4616. It has a TATA box (TATAAAAG) from -27 to -20 nts from the TSS.[7]
- Gene ID: 4744. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4842. It has a TATA box (TATAAAAG) from -27 to -20 nts from the TSS.[7]
- Gene ID: 5033. It has a TATA box (TATAAAAG) from -33 to -21 nts from the TSS.[7]
- Gene ID: 5054. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 5079. It has a TATA box (TATAAAAG) from -28 to -25 nts from the TSS.[7]
- Gene ID: 5225. It has a TATA box (TATAAAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 5406. It has a TATA box (TATAAAAG) from -24 to -21 nts from the TSS.[7]
- Gene ID: 5408. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 5478. It has a TATA box (TATAAAAG) from 8 to 15 nts from the TSS.[7]
- Gene ID: 5514. It has a TATA box (TATAAAAG) from -28 to -22 nts from the TSS.[7]
- Gene ID: 5741. It has a TATA box (TATAAAAG) from -29 to -21 nts from the TSS.[7]
- Gene ID: 6175. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 6187. It has a TATA box (TATAAAAG) from -25 to -18 nts from the TSS.[7]
- Gene ID: 6354. It has a TATA box (TATAAAAG) from -17 to -10 nts from the TSS.[7]
- Gene ID: 6356. It has a TATA box (TATAAAAG) from -44 to -37 nts from the TSS.[7]
- Gene ID: 6357. It has a TATA box (TATAAAAG) from -36 to -29 nts from the TSS.[7]
- Gene ID: 6698. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 6988. It has a TATA box (TATAAAAG) from -26 to -19 nts from the TSS.[7]
- Gene ID: 7038. It has a TATA box (TATAAAAG) from -35 to -28 nts from the TSS.[7]
- Gene ID: 7276. It has a TATA box (TATAAAAG) from -30 to -21 nts from the TSS.[7]
- Gene ID: 7280. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 7803. It has a TATA box (TATAAAAG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 7852. It has a TATA box (TATAAAAG) from -26 to -19 nts from the TSS.[7]
- Gene ID: 8288. It has a TATA box (TATAAAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 8339. It has a TATA box (TATAAAAG) from -33 to -26 nts from the TSS.[7]
- Gene ID: 8970. It has a TATA box (TATAAAAG) from -28 to -21 nts from the TSS.[7]
- Gene ID: 9510. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 9607. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 9709. It has a TATA box (TATAAAAG) from 1 to 8 nts from the TSS.[7]
- Gene ID: 11067. It has a TATA box (TATAAAAG) from 14 to 21 nts from the TSS.[7]
- Gene ID: 23645. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 27106. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 27106. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 50943. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 51297. It has a TATA box (TATAAAAG) from -45 to -38 nts from the TSS.[7]
- Gene ID: 56675. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 80177. It has a TATA box (TATAAAAG) from -34 to -27 nts from the TSS.[7]
- Gene ID: 81285. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 83998. It has a TATA box (TATAAAAG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 85235. It has a TATA box (TATAAAAG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 116842. It has a TATA box (TATAAAAG) from -32 to -25 nts from the TSS.[7]
TATA box (Basehoar 2004) samplings
For the Basic programs (starting with SuccessablesTATA.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction, looking for TATA(A/T)A(A/T)(A/G): 2, TATATAAA at 1602, TATATATA at 1600.
- Positive strand, negative direction: 3, TATATAAA at 2874, TATAAAAG at 223, TATAAAAG at 184.
- Negative strand, positive direction: 0.
- Positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction: 2, TTTATATA at 2871, TATATATA at 1600.
- inverse complement, positive strand, negative direction: 1, TTTTTATA at 219.
- inverse complement, negative strand, positive direction: 0.
- inverse complement, positive strand, positive direction: 0.
TATA (4560-2846) UTRs
- Negative strand, negative direction: TTTATATA at 2871.
- Positive strand, negative direction: TATATAAA at 2874.
TATA negative direction (2596-1) distal promoters
- Negative strand, negative direction: TATATAAA at 1602, TATATATA at 1600.
- Positive strand, negative direction: TATAAAAG at 223, TATAAAAG at 184.
- Positive strand, negative direction: TTTTTATA at 219.
TATA box (Basehoar 2004) random dataset samplings
- TATAr0: 0.
- TATAr1: 0.
- TATAr2: 0.
- TATAr3: 0.
- TATAr4: 0.
- TATAr5: 1, TATAAATG at 1565.
- TATAr6: 0.
- TATAr7: 1, TATAAATA at 3631.
- TATAr8: 1, TATAAAAA at 708.
- TATAr9: 1, TATATAAA at 3150.
- TATAr0ci: 0.
- TATAr1ci: 0.
- TATAr2ci: 0.
- TATAr3ci: 0.
- TATAr4ci: 2, CTTTTATA at 4527, CTTATATA at 4254.
- TATAr5ci: 0.
- TATAr6ci: 1, CTTATATA at 833.
- TATAr7ci: 1, TTTATATA at 4254.
- TATAr8ci: 0.
- TATAr9ci: 0.
TATAr arbitrary (evens) (4560-2846) UTRs
- TATAr4ci: CTTTTATA at 4527, CTTATATA at 4254.
TATAr alternate (odds) (4560-2846) UTRs
- TATAr7: TATAAATA at 3631.
- TATAr9: TATATAAA at 3150.
- TATAr7ci: TTTATATA at 4254.
TATAr arbitrary positive direction (odds) (4265-4050) proximal promoters
- TATAr7ci: TTTATATA at 4254.
TATAr alternate positive direction (evens) (4265-4050) proximal promoters
- TATAr4ci: CTTATATA at 4254.
TATAr arbitrary negative direction (evens) (2596-1) distal promoters
- TATAr8: TATAAAAA at 708.
- TATAr6ci: CTTATATA at 833.
TATAr alternate negative direction (odds) (2596-1) distal promoters
- TATAr5: TATAAATG at 1565.
TATAr arbitrary positive direction (odds) (4050-1) distal promoters
- TATAr5: TATAAATG at 1565.
- TATAr7: TATAAATA at 3631.
- TATAr9: TATATAAA at 3150.
TATAr alternate positive direction (evens) (4050-1) distal promoters
- TATAr8: TATAAAAA at 708.
- TATAr6ci: CTTATATA at 833.
TATA box (Basehoar 2004) analysis and results
"About 24% of human genes have a TATA-like element and their promoters are generally AT-rich; however, only ~10% of these TATA-containing promoters have the canonical TATA box (TATAWAWR)."[3] Several Saccharomyces genomes had the consensus sequence TATA(A/T)A(A/T)(A/G), yet only about 20% of yeast genes even contained the TATA sequence.[8]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 2 | 2 | 1 | 1 (--1,+-1) |
| Randoms | UTR | arbitrary negative | 2 | 10 | 0.2 | 0.25 |
| Randoms | UTR | alternate negative | 3 | 10 | 0.3 | 0.25 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0 |
| Reals | Proximal | negative | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 1 | 10 | 0.1 | 0 |
| Randoms | Proximal | alternate positive | 1 | 10 | 0.1 | 0 |
| Reals | Distal | negative | 5 | 2 | 2.5 | 2.5 ± 0.5 (--2,+-3) |
| Randoms | Distal | arbitrary negative | 2 | 10 | 0.2 | 0.15 |
| Randoms | Distal | alternate negative | 1 | 10 | 0.1 | 0.15 |
| Reals | Distal | positive | 0 | 2 | 0 | 0 |
| Randoms | Distal | arbitrary positive | 3 | 10 | 0.3 | 0.25 |
| Randoms | Distal | alternate positive | 2 | 10 | 0.2 | 0.25 |
Comparison:
The occurrences of real TATA UTRs and distals are greater than the randoms. This suggests that the real TATAs are likely active or activable.
Basehoar-1 TATA boxes
- Gene ID: 173. It has a TATA box (TATAAAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 229. It has a TATA box (TATAAAAA) from -32 to -24 nts from the TSS.[7]
- Gene ID: 229. It has a TATA box (TATAAAAA) from 23 to 30 nts from the TSS.[7]
- Gene ID: 759. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 1544. It has a TATA box (TATAAAAA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 1571. It has a TATA box (TATAAAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 1581. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 1906. It has a TATA box (TATAAAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 1938. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 1974. It has a TATA box (TATAAAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 2688. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 3605. It has a TATA box (TATAAAAA) from -44 to -37 nts from the TSS.[7]
- Gene ID: 4256. It has a TATA box (TATAAAAA) from 2 to 9 nts from the TSS.[7]
- Gene ID: 4435. It has a TATA box (TATAAAAA) from -2 to 6 nts from the TSS.[7]
- Gene ID: 4878. It has a TATA box (TATAAAAA) from -30 to -22 nts from the TSS.[7]
- Gene ID: 5743. It has a TATA box (TATAAAAA) from -31 to -25 nts from the TSS.[7]
- Gene ID: 6279. It has a TATA box (TATAAAAA) from -30 to -24 nts from the TSS.[7]
- Gene ID: 6376. It has a TATA box (TATAAAAA) from -12 to -5 nts from the TSS.[7]
- Gene ID: 6513. It has a TATA box (TATAAAAA) from 18 to 24 nts from the TSS.[7]
- Gene ID: 6518. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 6906. It has a TATA box (TATAAAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 8564. It has a TATA box (TATAAAAA) from -14 to -7 nts from the TSS.[7]
- Gene ID: 8832. It has a TATA box (TATAAAAA) from -12 to -5 nts from the TSS.[7]
- Gene ID: 8969. It has a TATA box (TATAAAAA) from -27 to -20 nts from the TSS.[7]
- Gene ID: 10215. It has a TATA box (TATAAAAA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 27159. It has a TATA box (TATAAAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 51129. It has a TATA box (TATAAAAA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 54106. It has a TATA box (TATAAAAA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 55603. It has a TATA box (TATAAAAA) from -2 to 6 nts from the TSS.[7]
- Gene ID: 64111. It has a TATA box (TATAAAAA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 130120. It has a TATA box (TATAAAAA) from 17 to 24 nts from the TSS.[7]
- Gene ID: 147183. It has a TATA box (TATAAAAA) from -30 to -23 nts from the TSS.[7]
Basehoar-2 TATA boxes
- Gene ID: 2. It has a TATA box (TATAAATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 183. It has a TATA box (TATAAATA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 203. It has a TATA box (TATAAATA) from -4 to 4 nts from the TSS.[7]
- Gene ID: 279. It has a TATA box (TATAAATA) from -27 to -20 nts from the TSS.[7]
- Gene ID: 280. It has a TATA box (TATAAATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 358. It has a TATA box (TATAAATA) from 23 to 30 nts from the TSS.[7]
- Gene ID: 358. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 846. It has a TATA box (TATAAATA) from -28 to -25 nts from the TSS.[7]
- Gene ID: 1051. It has a TATA box (TATAAATA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 1152. It has a TATA box (TATAAATA) from -41 to -34 nts from the TSS.[7]
- Gene ID: 1191. It has a TATA box (TATAAATA) from -23 to -16 nts from the TSS.[7]
- Gene ID: 1278. It has a TATA box (TATAAATA) from -34 to -27 nts from the TSS.[7]
- Gene ID: 2250. It has a TATA box (TATAAATA) from -32 to -21 nts from the TSS.[7]
- Gene ID: 3458. It has a TATA box (TATAAATA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 3906. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 4144. It has a TATA box (TATAAATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 4747. It has a TATA box (TATAAATA) from -28 to -21 nts from the TSS.[7]
- Gene ID: 4843. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 5449. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 6288. It has a TATA box (TATAAATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 6289. It has a TATA box (TATAAATA) from -17 to -10 nts from the TSS.[7]
- Gene ID: 6364. It has a TATA box (TATAAATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 6414. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
- Gene ID: 6428. It has a TATA box (TATAAATA) from 19 to 26 nts from the TSS.[7]
- Gene ID: 6707. It has a TATA box (TATAAATA) from -30 to -23 nts from the TSS.[7]
- Gene ID: 8431. It has a TATA box (TATAAATA) from -35 to -28 nts from the TSS.[7]
- Gene ID: 10563. It has a TATA box (TATAAATA) from 32 to 40 nts from the TSS.[7]
- Gene ID: 12723. It has a TATA box (TATAAATA) from -32 to -25 nts from the TSS.[7]
- Gene ID: 22928. It has a TATA box (TATAAATA) from -33 to -26 nts from the TSS.[7]
- Gene ID: 51582. It has a TATA box (TATAAATA) from -26 to -19 nts from the TSS.[7]
- Gene ID: 81606. It has a TATA box (TATAAATA) from -29 to -22 nts from the TSS.[7]
- Gene ID: 84223. It has a TATA box (TATAAATA) from 26 to 33 nts from the TSS.[7]
- Gene ID: 117158. It has a TATA box (TATAAATA) from -31 to -24 nts from the TSS.[7]
Basehoar-3 TATA boxes
- Gene ID: 338. It has a TATA box (TATAAATG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 359. It has a TATA box (TATAAATG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 383. It has a TATA box (TATAAATG) from -29 to -22 nts from the TSS.[7]
- Gene ID: 1427. It has a TATA box (TATAAATG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 4357. It has a TATA box (TATAAATG) from -17 to -10 nts from the TSS.[7]
- Gene ID: 4741. It has a TATA box (TATAAATG) from -32 to -25 nts from the TSS.[7]
- Gene ID: 6280. It has a TATA box (TATAAATG) from -23 to -16 nts from the TSS.[7]
- Gene ID: 6435. It has a TATA box (TATAAATG) from -42 to -35 nts from the TSS.[7]
- Gene ID: 6916. It has a TATA box (TATAAATG) from -30 to -23 nts from the TSS.[7]
- Gene ID: 10482. It has a TATA box (TATAAATG) from -31 to -24 nts from the TSS.[7]
- Gene ID: 51313. It has a TATA box (TATAAATG) from 2 to 9 nts from the TSS.[7]
- Gene ID: 126393. It has a TATA box (TATAAATG) from -30 to -23 nts from the TSS.[7]
M3 motif samplings
For the Basic programs testing consensus sequence (C/G)CGGAAG(C/T) (starting with SuccessablesM3.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction: 0.
- Positive strand, negative direction: 1, GCGGAAGT at 2731.
- Negative strand, positive direction: 0.
- Positive strand, positive direction: 7, CCGGAAGC at 1517, GCGGAAGC at 1306, CCGGAAGT at 1265, CCGGAAGC at 1013, CCGGAAGT at 929, CCGGAAGT at 829, CCGGAAGC at 593.
- inverse complement, negative strand, negative direction: 1, GCTTCCGT at 1558.
- inverse complement, positive strand, negative direction: 0.
- inverse complement, negative strand, positive direction: 0.
- inverse complement, positive strand, positive direction: 0.
M3 negative direction (2811-2596) proximal promoters
- Positive strand, negative direction: GCGGAAGT at 2731.
M3 negative direction (2596-1) distal promoters
- Negative strand, negative direction: GCTTCCGT at 1558.
M3 positive direction (4050-1) distal promoters
- Positive strand, positive direction: CCGGAAGC at 1517, GCGGAAGC at 1306, CCGGAAGT at 1265, CCGGAAGC at 1013, CCGGAAGT at 929, CCGGAAGT at 829, CCGGAAGC at 593.
M3 random dataset samplings
- M3r0: 0.
- M3r1: 0.
- M3r2: 1, GCGGAAGT at 3374.
- M3r3: 0.
- M3r4: 1, GCGGAAGT at 2555.
- M3r5: 0.
- M3r6: 2, GCGGAAGC at 2580, CCGGAAGT at 1193.
- M3r7: 0.
- M3r8: 1, GCGGAAGC at 2757.
- M3r9: 0.
- M3r0ci: 0.
- M3r1ci: 0.
- M3r2ci: 1, ACTTCCGG at 3397.
- M3r3ci: 1, GCTTCCGC at 1824.
- M3r4ci: 0.
- M3r5ci: 0.
- M3r6ci: 0.
- M3r7ci: 0.
- M3r8ci: 1, ACTTCCGC at 4254.
- M3r9ci: 1, GCTTCCGG at 3340.
M3r arbitrary (evens) (4560-2846) UTRs
- M3r2: GCGGAAGT at 3374.
- M3r2ci: ACTTCCGG at 3397.
- M3r8ci: ACTTCCGC at 4254.
M3r alternate (odds) (4560-2846) UTRs
- M3r9ci: GCTTCCGG at 3340.
M3r arbitrary negative direction (evens) (2811-2596) proximal promoters
- M3r8: GCGGAAGC at 2757.
M3r alternate positive direction (evens) (4265-4050) proximal promoters
- M3r8ci: ACTTCCGC at 4254.
M3r arbitrary negative direction (evens) (2596-1) distal promoters
- M3r4: GCGGAAGT at 2555.
- M3r6: GCGGAAGC at 2580, CCGGAAGT at 1193.
M3r alternate negative direction (odds) (2596-1) distal promoters
- M3r3ci: GCTTCCGC at 1824.
M3r arbitrary positive direction (odds) (4050-1) distal promoters
- M3r3ci: GCTTCCGC at 1824.
- M3r9ci: GCTTCCGG at 3340.
M3r alternate positive direction (evens) (4050-1) distal promoters
- M3r2: GCGGAAGT at 3374.
- M3r4: GCGGAAGT at 2555.
- M3r6: GCGGAAGC at 2580, CCGGAAGT at 1193.
- M3r8: GCGGAAGC at 2757.
- M3r2ci: ACTTCCGG at 3397.
M3 analysis and results
M3 (SCGGAAGY) occurs preferentially in human TATA-less core promoters.[3]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 0 | 2 | 0 | 0 |
| Randoms | UTR | arbitrary negative | 3 | 10 | 0.3 | 0.2 ± 0.1 |
| Randoms | UTR | alternate negative | 1 | 10 | 0.1 | 0.2 ± 0.1 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0 |
| Reals | Proximal | negative | 1 | 2 | 0.5 | 0.5 ± 0.5 (--0,+-1) |
| Randoms | Proximal | arbitrary negative | 1 | 10 | 0.1 | 0.05 |
| Randoms | Proximal | alternate negative | 0 | 10 | 0 | 0.05 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 0 | 10 | 0 | 0.05 |
| Randoms | Proximal | alternate positive | 1 | 10 | 0.1 | 0.05 |
| Reals | Distal | negative | 1 | 2 | 0.5 | 0.5 ± (--1,+-0) |
| Randoms | Distal | arbitrary negative | 3 | 10 | 0.3 | 0.2 ± 0.1 |
| Randoms | Distal | alternate negative | 1 | 10 | 0.1 | 0.2 ± 0.1 |
| Reals | Distal | positive | 7 | 2 | 3.5 | 3.5 ± 3.5 (-+0,++7) |
| Randoms | Distal | arbitrary positive | 2 | 10 | 0.2 | 0.4 ± 0.2 |
| Randoms | Distal | alternate positive | 6 | 10 | 0.6 | 0.4 ± 0.2 |
Comparison:
The occurrences of real M3 proximal and distals are greater than the randoms. This suggests that the real M3s are likely active or activable.
M22 samplings
For the Basic programs testing consensus sequence TGCGCAN(G/T) (starting with SuccessablesM22.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are, are looking for, and found:
- Negative strand, negative direction: 0.
- Positive strand, negative direction: 0.
- Negative strand, positive direction: 1, TGCGCAAG at 1525.
- Positive strand, positive direction: 0.
- inverse complement, negative strand, negative direction: 0.
- inverse complement, positive strand, negative direction: 0.
- inverse complement, negative strand, positive direction: 3, CGTGCGCA at 1523, CCTGCGCA at 1414, CCTGCGCA at 1314.
- inverse complement, positive strand, positive direction: 0.
M22 positive direction (4050-1) distal promoters
- Negative strand, positive direction: TGCGCAAG at 1525.
- Negative strand, positive direction: CGTGCGCA at 1523, CCTGCGCA at 1414, CCTGCGCA at 1314.
M22 random dataset samplings
- M22r0: 0.
- M22r1: 1, TGCGCAAT at 1589.
- M22r2: 0.
- M22r3: 0.
- M22r4: 0.
- M22r5: 0.
- M22r6: 0.
- M22r7: 0.
- M22r8: 0.
- M22r9: 0.
- M22r0ci: 0.
- M22r1ci: 0.
- M22r2ci: 0.
- M22r3ci: 0.
- M22r4ci: 0.
- M22r5ci: 0.
- M22r6ci: 0.
- M22r7ci: 0.
- M22r8ci: 0.
- M22r9ci: 0.
M22r alternate negative direction (odds) (2596-1) distal promoters
- M22r1: TGCGCAAT at 1589.
M22r arbitrary positive direction (odds) (4050-1) distal promoters
- M22r1: TGCGCAAT at 1589.
M22 analysis and results
M22 (TGCGCANK), where K = (G/T) occurs preferentially in human TATA-less core promoters.[3]
| Reals or randoms | Promoters | direction | Numbers | Strands | Occurrences | Averages (± 0.1) |
|---|---|---|---|---|---|---|
| Reals | UTR | negative | 0 | 2 | 0 | 0 |
| Randoms | UTR | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | UTR | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | negative | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Core | positive | 0 | 2 | 0 | 0 |
| Randoms | Core | arbitrary positive | 0 | 10 | 0 | 0 |
| Randoms | Core | alternate positive | 0 | 10 | 0 | 0 |
| Reals | Proximal | negative | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary negative | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate negative | 0 | 10 | 0 | 0 |
| Reals | Proximal | positive | 0 | 2 | 0 | 0 |
| Randoms | Proximal | arbitrary positive | 0 | 10 | 0 | 0 |
| Randoms | Proximal | alternate positive | 0 | 10 | 0 | 0 |
| Reals | Distal | negative | 0 | 2 | 0 | 0 |
| Randoms | Distal | arbitrary negative | 0 | 10 | 0 | 0.05 |
| Randoms | Distal | alternate negative | 1 | 10 | 0.1 | 0.05 |
| Reals | Distal | positive | 4 | 2 | 2 | 2 ± 2 (-+4,++0) |
| Randoms | Distal | arbitrary positive | 1 | 10 | 0.1 | 0.05 |
| Randoms | Distal | alternate positive | 0 | 10 | 0 | 0.05 |
Comparison:
The occurrences of real M22 distals are greater than the randoms. This suggests that the real M22s are likely active or activable.
Comparisons of TATA boxes for UTRs nn
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (4560-2846) | (4560-2846) | (4560-2846) | (4560-2846) | (4560-2846) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| - | - | TATATAT at 2872 | - | - |
| - | - | ciTTATATA at 2871 | ciTTTATATA at 2871 | ciTTTATATA at 2871 |
| ciTTTATA at 2869 | ciTTTTATA at 2869 | ciTTTTATA at 2869 | - | - |
| TATAAA at 2852 | TATAAAA at 2853 | TATAAAA at 2853 | - | - |
Comparisons of TATA boxes for UTRs pn
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (4560-2846) | (4560-2846) | (4560-2846) | (4560-2846) | (4560-2846) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| TATAAA at 2874 | - | TATATAA at 2873 | TATATAAA at 2874 | TATATAAA at 2874 |
Comparisons of TATA boxes for proximals nn
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (2811-2596) | (2811-2596) | (2811-2596) | (2811-2596) | (2811-2596) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| ciTTTATA at 2638 | - | ciATTTATA at 2638 | - | - |
Comparisons of TATA boxes for distals nn
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (2596-1) | (2596-1) | (2596-1) | (2596-1) | (2596-1) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| ciTTTATA at 1740 | ciTTTTATA at 1740 | ciTTTTATA at 1740 | - | - |
| TATAAA at 1602 | - | TATATAA at 1601 | TATATAAA at 1602 | TATATAAA at 1602 |
| - | - | TATATAT at 1599 | - | TATATATA at 1600 |
Comparisons of TATA boxes for distals pn
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (2596-1) | (2596-1) | (2596-1) | (2596-1) | (2596-1) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| - | - | TATATAT at 1600 | - | - |
| TATAAA at 221 | TATAAAA at 222 | TATAAAA at 222 | TATAAAAG at 223 | TATAAAAG at 223 |
| ciTTTATA at 219 | ciTTTTATA at 219 | ciTTTTATA at 219 | ciTTTTTATA at 219 | ciTTTTTATA at 219 |
| TATAAA at 182 | TATAAAA at 183 | TATAAAA at 183 | TATAAAAG at 184 | ciTATAAAAG at 184 |
Comparisons of TATA boxes for distals np
| Butler (2002) | Carninci (2006) | Watson (2014) | Juven-Gershon (2008) | Basehoar (2004) |
|---|---|---|---|---|
| (4050-1) | (4050-1) | (4050-1) | (4050-1) | (4050-1) |
| TATAAA | TATAAAA | TATA(A/T)A(A/T) | TATA(A/T)AA(A/G) | TATA(A/T)A(A/T)(A/G) |
| ciTTTATA at 2588 | - | - | - | - |
Acknowledgements
The content on this page was first contributed by: Henry A. Hoff.
Initial content for this page in some instances came from Wikiversity.
See also
References
- ↑ R. P. Lifton, M. L. Goldberg, R. W. Karp, and D. S. Hogness (1978). "The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications". Cold Spring Harbor Symposia on Quantitative Biology. 42: 1047–51. doi:10.1101/SQB.1978.042.01.105. PMID 98262.
- ↑ 2.0 2.1 Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter" (PDF). Annual Review of Biochemistry. 72 (1): 449–79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. Retrieved 2012-05-07.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 C Yang, E Bolotin, T Jiang, FM Sladek, E Martinez (March 2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene. 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMID 17123746.
- ↑ Stephen T. Smale (October 1, 2001). "Core promoters: active contributors to combinatorial gene regulation". Genes & Development. 15 (19): 2503–8. doi:10.1101/gad.937701. Retrieved 2012-04-28.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 Tamar Juven-Gershon and James T. Kadonaga (15 March 2010). "Regulation of gene expression via the core promoter and the basal transcriptional machinery". Developmental Biology. 339 (2): 225–9. doi:10.1016/j.ydbio.2009.08.009. Retrieved 2016-01-16.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Muyu Xu, Elsie Gonzalez-Hurtado, and Ernest Martinez (April 2016). "Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1859 (4): 553–563. doi:10.1016/j.bbagrm.2016.01.005. Retrieved 2024-06-08.
- ↑ 7.000 7.001 7.002 7.003 7.004 7.005 7.006 7.007 7.008 7.009 7.010 7.011 7.012 7.013 7.014 7.015 7.016 7.017 7.018 7.019 7.020 7.021 7.022 7.023 7.024 7.025 7.026 7.027 7.028 7.029 7.030 7.031 7.032 7.033 7.034 7.035 7.036 7.037 7.038 7.039 7.040 7.041 7.042 7.043 7.044 7.045 7.046 7.047 7.048 7.049 7.050 7.051 7.052 7.053 7.054 7.055 7.056 7.057 7.058 7.059 7.060 7.061 7.062 7.063 7.064 7.065 7.066 7.067 7.068 7.069 7.070 7.071 7.072 7.073 7.074 7.075 7.076 7.077 7.078 7.079 7.080 7.081 7.082 7.083 7.084 7.085 7.086 7.087 7.088 7.089 7.090 7.091 7.092 7.093 7.094 7.095 7.096 7.097 7.098 7.099 7.100 7.101 7.102 7.103 7.104 7.105 7.106 7.107 7.108 7.109 7.110 7.111 7.112 7.113 7.114 7.115 7.116 7.117 7.118 7.119 7.120 7.121 7.122 7.123 7.124 7.125 7.126 7.127 7.128 7.129 7.130 7.131 7.132 7.133 7.134 7.135 7.136 7.137 7.138 7.139 7.140 7.141 7.142 7.143 7.144 7.145 7.146 7.147 7.148 7.149 7.150 7.151 7.152 7.153 7.154 7.155 7.156 7.157 7.158 7.159 7.160 7.161 7.162 7.163 7.164 7.165 7.166 7.167 7.168 7.169 7.170 7.171 7.172 7.173 7.174 7.175 7.176 7.177 7.178 7.179 7.180 7.181 7.182 7.183 7.184 7.185 7.186 7.187 7.188 7.189 7.190 7.191 7.192 7.193 7.194 7.195 7.196 7.197 7.198 7.199 7.200 7.201 7.202 7.203 7.204 7.205 7.206 7.207 7.208 7.209 7.210 7.211 7.212 7.213 7.214 7.215 7.216 7.217 7.218 7.219 7.220 7.221 7.222 7.223 7.224 7.225 7.226 7.227 7.228 7.229 7.230 7.231 7.232 7.233 7.234 7.235 7.236 7.237 7.238 7.239 7.240 7.241 7.242 7.243 7.244 7.245 7.246 7.247 7.248 7.249 7.250 7.251 7.252 7.253 7.254 7.255 7.256 7.257 7.258 7.259 7.260 7.261 7.262 7.263 7.264 7.265 7.266 7.267 7.268 7.269 7.270 7.271 7.272 7.273 7.274 7.275 7.276 7.277 7.278 7.279 7.280 7.281 7.282 7.283 7.284 7.285 7.286 7.287 7.288 7.289 7.290 7.291 7.292 7.293 7.294 7.295 7.296 7.297 7.298 7.299 7.300 7.301 7.302 7.303 7.304 7.305 7.306 7.307 7.308 7.309 7.310 7.311 7.312 7.313 7.314 7.315 7.316 7.317 7.318 7.319 7.320 7.321 7.322 7.323 7.324 7.325 7.326 7.327 7.328 7.329 7.330 7.331 7.332 7.333 7.334 7.335 7.336 7.337 7.338 7.339 7.340 7.341 7.342 7.343 7.344 7.345 7.346 7.347 7.348 7.349 7.350 7.351 7.352 7.353 7.354 7.355 7.356 7.357 7.358 7.359 7.360 7.361 7.362 7.363 7.364 7.365 7.366 7.367 7.368 7.369 7.370 7.371 7.372 7.373 7.374 7.375 7.376 7.377 7.378 7.379 7.380 7.381 7.382 7.383 7.384 7.385 7.386 7.387 7.388 7.389 7.390 7.391 7.392 7.393 7.394 7.395 7.396 7.397 7.398 7.399 7.400 7.401 7.402 7.403 7.404 7.405 7.406 7.407 7.408 7.409 7.410 7.411 7.412 7.413 7.414 7.415 7.416 7.417 7.418 7.419 7.420 7.421 7.422 7.423 7.424 7.425 7.426 7.427 7.428 7.429 7.430 7.431 7.432 7.433 7.434 7.435 7.436 7.437 7.438 7.439 7.440 7.441 7.442 7.443 7.444 7.445 7.446 7.447 7.448 7.449 7.450 7.451 7.452 7.453 7.454 7.455 7.456 7.457 7.458 7.459 7.460 7.461 7.462 7.463 7.464 7.465 7.466 7.467 7.468 7.469 7.470 7.471 7.472 7.473 7.474 7.475 7.476 7.477 7.478 7.479 7.480 7.481 7.482 7.483 7.484 7.485 7.486 7.487 7.488 7.489 7.490 7.491 7.492 7.493 7.494 7.495 7.496 7.497 7.498 7.499 7.500 7.501 7.502 7.503 7.504 7.505 7.506 7.507 7.508 7.509 7.510 7.511 7.512 7.513 7.514 7.515 7.516 7.517 7.518 7.519 7.520 7.521 7.522 7.523 7.524 7.525 7.526 7.527 7.528 7.529 7.530 7.531 7.532 7.533 7.534 7.535 7.536 7.537 7.538 7.539 7.540 7.541 7.542 7.543 7.544 7.545 7.546 7.547 7.548 7.549 7.550 7.551 7.552 7.553 7.554 7.555 7.556 7.557 7.558 7.559 7.560 7.561 7.562 7.563 7.564 7.565 7.566 7.567 7.568 7.569 7.570 7.571 7.572 7.573 7.574 7.575 7.576 7.577 7.578 7.579 7.580 7.581 7.582 7.583 7.584 7.585 7.586 7.587 7.588 7.589 7.590 7.591 7.592 7.593 Victor X Jin, Gregory AC Singer, Francisco J Agosto-Pérez, Sandya Liyanarachchi, and Ramana V Davuluri (2006). "Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs". BMC Bioinformatics. 7: 114. doi:10.1186/1471-2105-7-114. Retrieved 2024-06-09.
- ↑ 8.0 8.1 Basehoar, Andrew D.; Zanton, Sara J.; Pugh, B. Franklin (2004-03-05). "Identification and distinct regulation of yeast TATA box-containing genes". Cell. 116 (5): 699–709. ISSN 0092-8674. PMID 15006352.
- ↑ Jennifer E.F. Butler, James T. Kadonaga (October 15, 2002). "The RNA polymerase II core promoter: a key component in the regulation of gene expression". Genes & Development. 16 (20): 2583–292. doi:10.1101/gad.1026202. PMID 12381658.
- ↑ Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engström P.G., Frith M.C., Forrest A.R., Alkema W.B., Tan S.L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S.M., Wells C.A., Orlando V., Wahlestedt C., Liu E.T., Harbers M., Kawai J., Bajic V.B., Hume D.A., Hayashizaki Y. (2006). "Genome-wide analysis of mammalian promoter architecture and evolution". Nat. Genet. 38 (6): 626–35. doi:10.1038/ng1789. PMID 16645617.
- ↑ Molecular biology of the gene. Watson, James D., 1928- (Seventh ed.). Boston. ISBN 9780321762436. OCLC 824087979.
Further reading
- C Yang, E Bolotin, T Jiang, FM Sladek, E Martinez (March 2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene. 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMC 1955227. PMID 17123746.
External links
- GenomeNet KEGG database
- Home - Gene - NCBI
- NCBI All Databases Search
- NCBI Site Search
- PubChem Public Chemical Database