Systemic lupus erythematosus pathophysiology: Difference between revisions
Usama Talib (talk | contribs) |
|||
| (13 intermediate revisions by 3 users not shown) | |||
| Line 9: | Line 9: | ||
{{Systemic lupus erythematosus}} | {{Systemic lupus erythematosus}} | ||
{{CMG}}; {{AE}} {{MIR}} {{CZ}} {{RT}} | {{CMG}}; {{AE}} {{MIR}}, {{CZ}}, {{RT}} | ||
==Overview== | ==Overview== | ||
The pathophysiology of systemic lupus erythematosus involves the [[immune system]]. Other factors such as [[genetic]] factors, [[hormonal]] abnormalities, and environmental factors also play a role. The most important environmental factors involved in the [[pathogenesis]] of SLE include [[ultraviolet]] (UV) light and some [[infections]]. The most important [[Gene|genes]] involved in the [[pathogenesis]] of SLE include [[HLA-DR2]], [[HLA-DR3]], [[HLA]] class 3, C1q, and [[IRF5|interferon (IFN) regulatory factor 5]]. The most prominent events involving [[immune]] abnormalities are related to persistent activation of [[B cells]] and [[Plasma cell|plasma cells]] that make [[Autoantibody|auto-antibodies]] during disease progression. The disease developmental process begins with the release of microparticles and [[proinflammatory]] [[cytokines]] from the cells that are undergoing [[apoptosis]]. Due to excess amount of [[apoptosis]], the body is unable to clear these microparticles entirely, and these microparticles are presented to [[dendritic cells]] as [[antigens]]. [[Dendritic cells]] process these microparticles and mature, and present these as [[antigens]] to [[T-cells]]. [[T-cells]], microparticles, and [[proinflammatory]] [[cytokines]] themselves trigger [[B-cell]] activation and [[autoantibody]] production. As a result, body tissues lose their self-tolerance. The most prominent events involving [[hormonal]] abnormalities are due to [[prolactin]] and [[estrogen]]. On microscopic [[histopathological]] analysis, [[apoptotic]] [[Keratinocyte|keratinocytes]], [[vacuolization]] of the [[basement membrane]], and [[dermal]] [[mucin]] deposition are characteristic findings of SLE [[dermatitis]], and active or inactive [[Endocapillary proliferative glomerulonephritis|endocapillary]] or extracapillary segmental [[glomerulonephritis]] are characteristic findings of [[lupus nephritis]]. | The pathophysiology of systemic lupus erythematosus involves the [[immune system]]. Other factors such as [[genetic]] factors, [[hormonal]] abnormalities, and environmental factors also play a role. The most important environmental factors involved in the [[pathogenesis]] of SLE include [[ultraviolet]] (UV) light and some [[infections]]. The most important [[Gene|genes]] involved in the [[pathogenesis]] of SLE include [[HLA-DR2]], [[HLA-DR3]], [[HLA]] class 3, C1q, and [[IRF5|interferon (IFN) regulatory factor 5]]. The most prominent events involving [[immune]] abnormalities are related to persistent activation of [[B cells]] and [[Plasma cell|plasma cells]] that make [[Autoantibody|auto-antibodies]] during disease progression. The disease developmental process begins with the release of microparticles and [[proinflammatory]] [[cytokines]] from the cells that are undergoing [[apoptosis]]. Due to excess amount of [[apoptosis]], the body is unable to clear these microparticles entirely, and these microparticles are presented to [[dendritic cells]] as [[antigens]]. [[Dendritic cells]] process these microparticles and mature, and present these as [[antigens]] to [[T-cells]]. [[T-cells]], microparticles, and [[proinflammatory]] [[cytokines]] themselves trigger [[B-cell]] activation and [[autoantibody]] production. As a result, body tissues lose their self-tolerance. The most prominent events involving [[hormonal]] abnormalities are due to [[prolactin]] and [[estrogen]]. On microscopic [[histopathological]] analysis, [[apoptotic]] [[Keratinocyte|keratinocytes]], [[vacuolization]] of the [[basement membrane]], and [[dermal]] [[mucin]] deposition are characteristic findings of SLE [[dermatitis]], and active or inactive [[Endocapillary proliferative glomerulonephritis|endocapillary]] or extracapillary segmental [[glomerulonephritis]] are characteristic findings of [[lupus nephritis]]. | ||
==Pathogenesis== | ==Pathogenesis== | ||
The progression of systemic lupus erythematosus (SLE) involves the [[immune system]]. Nearly all of the [[pathological]] manifestations of SLE | The progression of systemic lupus erythematosus (SLE) involves the [[immune system]]. Nearly all of the [[pathological]] manifestations of SLE occur due to [[antibody]] formation and the creation and deposition of [[immune complexes]] in different organs of the body. When the [[immune complexes]] are formed, they deposit on different body tissues and [[vessels]], which may lead to [[complement]] activation and more organ damage. There are other factors such as [[genetic]] factors, hormonal abnormalities, and environmental factors that also play a role in the pathogenesis of SLE. | ||
[[File:Lupus2.jpg|left|size:800×591 pixels]] | [[File:Lupus2.jpg|left|size:800×591 pixels]] | ||
<br clear="left" /> | <br clear="left" /> | ||
=== Environmental factors === | === Environmental factors === | ||
The environmental factors and genetic factors are the most important risk factors for developing SLE | The environmental factors and genetic factors are the most important risk factors for developing SLE because they may jump-start the disinhibited cellular [[apoptosis]] chain. This [[apoptosis]] step is the first step in the pathogenesis of lupus. | ||
* Infections | * Infections | ||
** May stimulate some [[antigen]] specific cells and increase [[apoptosis]] | ** May stimulate some [[antigen]] specific cells and increase [[apoptosis]] | ||
| Line 36: | Line 36: | ||
=== Immune abnormalities === | === Immune abnormalities === | ||
The development of systemic lupus erythematosus (SLE) is due to the activation of different mechanisms that may result in [[auto-immune|auto-immunity]]. The disease developmental process begins with the release of microparticles and [[proinflammatory]] [[cytokines]] from the cells that are undergoing [[apoptosis]]. Due to excess amount of [[apoptosis]], the body is unable to clear these microparticles entirely, and these microparticles are presented to [[dendritic cells]] as [[antigens]]. [[Dendritic cells]] process these microparticles and mature, and present these as [[antigens]] to [[T-cells]]. [[T-cells]], microparticles, and [[proinflammatory]] [[cytokines]] themselves trigger [[B-cell]] activation and [[autoantibody]] production. As a result, body tissues lose their self-tolerance. Affected patients are no longer entirely tolerant to all of their [[Antigens|self-antigens]], leading to development of an [[Autoimmunity|autoimmune]] disease and producing [[autoantibodies]] as a response. During disease progression, [[B cell|B cells]] and [[Plasma cell|plasma cells]] that make [[Autoantibody|autoantibodies]] are more persistently activated due to signaling abnormalities, causing them to make more [[Autoantibody|autoantibodies]]. These [[autoantibodies]] are targeted predominantly to [[intracellular]] [[nucleoprotein]] particles.<ref name="pmid8519610">{{cite journal |vauthors=Elkon K |title=Autoantibodies in systemic lupus erythematosus |journal=Curr Opin Rheumatol |volume=7 |issue=5 |pages=384–8 |year=1995 |pmid=8519610 |doi= |url=}}</ref><ref name="pmid25449682">{{cite journal |vauthors=Yaniv G, Twig G, Shor DB, Furer A, Sherer Y, Mozes O, Komisar O, Slonimsky E, Klang E, Lotan E, Welt M, Marai I, Shina A, Amital H, Shoenfeld Y |title=A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients |journal=Autoimmun Rev |volume=14 |issue=1 |pages=75–9 |year=2015 |pmid=25449682 |doi=10.1016/j.autrev.2014.10.003 |url=}}</ref> This increase in [[autoantibody]] production and persistence is supposed to be [[Downregulate|downregulated]] by anti-idiotypic [[antibodies]] or regulatory [[immune cells]], but the massive [[Immunology|immunologic]] response in SLE prevents this [[downregulation]] from taking place. After formation of [[immune complexes]], the [[classical complement pathway]] is activated, which leads to the deposition of [[immune complexes]] in different organs and is responsible for flare ups and long term complications. The most important [[immune]] abnormalities that are related to SLE development and progression are: | |||
==== Microparticles ==== | ==== Microparticles ==== | ||
| Line 51: | Line 51: | ||
==== Signaling abnormalities ==== | ==== Signaling abnormalities ==== | ||
[[Protein kinase|Protein kinases]] are responsible for [[intracellular]] [[cytokine]] signals. [[Intracellular signaling]] | [[Protein kinase|Protein kinases]] are responsible for [[intracellular]] [[cytokine]] signals. [[Intracellular signaling]] leads to various types of cell response, such as: | ||
* [[Cell migration]] | * [[Cell migration]] | ||
* [[Cell proliferation]] | * [[Cell proliferation]] | ||
* [[Inflammatory response]] | * [[Inflammatory response]] | ||
[[Cell signaling]] abnormalities | [[Cell signaling]] abnormalities leads to: | ||
* T and B [[Lymphocyte|lymphocytes]] cellular hyperactivity | * T and B [[Lymphocyte|lymphocytes]] cellular hyperactivity | ||
* T and B [[Lymphocyte|lymphocytes]] hyper responsiveness | * T and B [[Lymphocyte|lymphocytes]] hyper responsiveness | ||
* Persistence of | * Persistence of auto-reactive [[T cell|T cells]] that would otherwise have been deleted | ||
Signaling abnormalities of T and B [[Lymphocyte|lymphocytes]], may be due to: | Signaling abnormalities of T and B [[Lymphocyte|lymphocytes]], may be due to: | ||
* | * Abnormal [[Potassium channels|voltage-gated potassium channels]], these channels facilitate excessive [[calcium]] entry into [[T cells]] and lead to increased [[calcium]] responses to [[antigen]] stimulation | ||
* [[Hyperphosphorylation]] of [[cytosolic]] [[protein]] substrates | * [[Hyperphosphorylation]] of [[cytosolic]] [[protein]] substrates | ||
* Decreased nuclear factor kB | * Decreased nuclear factor kB | ||
==== B-Cell role ==== | ==== B-Cell role ==== | ||
* Increase in circulating [[plasma cells]] and [[Memory B cell|memory B cells]] that are associated with SLE activity | * Increase in circulating [[plasma cells]] and [[Memory B cell|memory B cells]] that are associated with SLE activity | ||
| Line 71: | Line 70: | ||
* Decrease in [[cytotoxic T cells]], decrease in [[suppressor T cells|suppressor T cell]] function, and impaired generation of [[T-cell|polyclonal T-cell]] [[cytolytic]] activity | * Decrease in [[cytotoxic T cells]], decrease in [[suppressor T cells|suppressor T cell]] function, and impaired generation of [[T-cell|polyclonal T-cell]] [[cytolytic]] activity | ||
* Increased number and activity of [[T helper cell|helper T cells]] | * Increased number and activity of [[T helper cell|helper T cells]] | ||
===== Neutrophil role ===== | ===== Neutrophil role ===== | ||
*Increased number of circulating [[neutrophils]] undergoing NETosis (NET=[[neutrophil extracellular traps]]), a form of [[apoptosis]] specific for [[Neutrophil|neutrophils]], releases [[DNA]] bound to [[protein]] in protein nets, which stimulates anti-DNA and [[Interferon-alpha|IFN-alpha]] production | *Increased number of circulating [[neutrophils]] undergoing NETosis (NET=[[neutrophil extracellular traps]]), a form of [[apoptosis]] specific for [[Neutrophil|neutrophils]], releases [[DNA]] bound to [[protein]] in protein nets, which stimulates anti-DNA and [[Interferon-alpha|IFN-alpha]] production | ||
| Line 84: | Line 79: | ||
=== Hormonal abnormalities === | === Hormonal abnormalities === | ||
The following evidence is suggestive of the [[Hormone|hormonal]] predisposition to SLE: | The following evidence is suggestive of the [[Hormone|hormonal]] predisposition to SLE: | ||
* | * Predilection of the disease for females shows the relationship between female hormones and the onset of SLE | ||
* Significantly increased risk for SLE in:<ref name="pmid17393454">{{cite journal |vauthors=Costenbader KH, Feskanich D, Stampfer MJ, Karlson EW |title=Reproductive and menopausal factors and risk of systemic lupus erythematosus in women |journal=Arthritis Rheum. |volume=56 |issue=4 |pages=1251–62 |year=2007 |pmid=17393454 |doi=10.1002/art.22510 |url=}}</ref> | * Significantly increased risk for SLE in:<ref name="pmid17393454">{{cite journal |vauthors=Costenbader KH, Feskanich D, Stampfer MJ, Karlson EW |title=Reproductive and menopausal factors and risk of systemic lupus erythematosus in women |journal=Arthritis Rheum. |volume=56 |issue=4 |pages=1251–62 |year=2007 |pmid=17393454 |doi=10.1002/art.22510 |url=}}</ref> | ||
** Early age of [[menarche]] | ** Early age of [[menarche]] | ||
| Line 104: | Line 99: | ||
*May inhibit the type 1 [[interferon]] pathway, suggesting that a balance between [[estrogen]] and [[progesterone]] may be critical for the body to remain healthy | *May inhibit the type 1 [[interferon]] pathway, suggesting that a balance between [[estrogen]] and [[progesterone]] may be critical for the body to remain healthy | ||
** Down-regulates [[T cell|T-cell]] proliferation and increases the number of [[CD8 cytotoxic lymphocyte|CD8 cells]] | ** Down-regulates [[T cell|T-cell]] proliferation and increases the number of [[CD8 cytotoxic lymphocyte|CD8 cells]] | ||
* Both [[progesterone]] and high levels of [[estrogen]] promote a [[Th2]] response, which favors [[autoantibody]] production | ** Act mainly as a protective agent | ||
* Both [[progesterone]] and high levels of [[estrogen]] promote a [[Th2]] response, which favors [[autoantibody|auto-antibody]] production | |||
==Genetics== | ==Genetics== | ||
Systemic lupus erythematosus is transmitted in [[polygenic inheritance]] pattern. [[Genes]] involved in the [[pathogenesis]] of systemic lupus erythematosus include [[HLA]] class 2 especially DR2 | Systemic lupus erythematosus is transmitted in a [[polygenic inheritance]] pattern. [[Genes]] involved in the [[pathogenesis]] of systemic lupus erythematosus include [[HLA]] class 2 (especially DR2 and DR3), [[HLA]] class 3 (especially complement genes including C2 and C4 genes), IFNRF5 gene, and other genes related to the [[Immune systems|immunologic system]]. | ||
The following evidence is also suggestive of the [[genetic predisposition]] of SLE:<ref name="pmid10768211">{{cite journal |vauthors=Sullivan KE |title=Genetics of systemic lupus erythematosus. Clinical implications |journal=Rheum. Dis. Clin. North Am. |volume=26 |issue=2 |pages=229–56, v–vi |year=2000 |pmid=10768211 |doi= |url=}}</ref> | The following evidence is also suggestive of the [[genetic predisposition]] of SLE:<ref name="pmid10768211">{{cite journal |vauthors=Sullivan KE |title=Genetics of systemic lupus erythematosus. Clinical implications |journal=Rheum. Dis. Clin. North Am. |volume=26 |issue=2 |pages=229–56, v–vi |year=2000 |pmid=10768211 |doi= |url=}}</ref> | ||
* Increase occurrence | * Increase occurrence of disease in [[identical twins]] | ||
* Increased disease frequency among first degree relatives | * Increased disease frequency among first degree relatives | ||
* The increased occurrence of the disease in siblings of SLE patients | * The increased occurrence of the disease in siblings of SLE patients | ||
{| class="wikitable" | {| class="wikitable" | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Class | ||
! style="background: #4479BA; color: #FFFFFF; " align="center" |Gene subtype | |||
! colspan="1" rowspan="1 | ! colspan="1" rowspan="1" style="background: #4479BA; color: #FFFFFF; " align="center" |Function | ||
! style="background: #4479BA; color: #FFFFFF; " align="center" |Pathological effect and Molecular mechanisms | |||
|- | |- | ||
! style="background:#DCDCDC;" |Autoantigen presentation | ! style="background:#DCDCDC;" |Autoantigen presentation | ||
| | | style="background:#DCDCDC;" align="center" |[[HLA]] class 2<ref name="pmid12867584">{{cite journal |vauthors=Lee HS, Chung YH, Kim TG, Kim TH, Jun JB, Jung S, Bae SC, Yoo DH |title=Independent association of HLA-DR and FCgamma receptor polymorphisms in Korean patients with systemic lupus erythematosus |journal=Rheumatology (Oxford) |volume=42 |issue=12 |pages=1501–7 |year=2003 |pmid=12867584 |doi=10.1093/rheumatology/keg404 |url=}}</ref> | ||
| colspan="1" rowspan="1" | | | colspan="1" rowspan="1" | | ||
* Contains [[genes]] encoding [[glycoproteins]] that process and present [[peptides]] for recognition by [[T cells]] ([[Antigen-presenting cell|antigen presenting cells]]) | * Contains [[genes]] encoding [[glycoproteins]] that process and present [[peptides]] for recognition by [[T cells]] ([[Antigen-presenting cell|antigen presenting cells]]) | ||
| Line 126: | Line 122: | ||
** [[HLA-DR3]] | ** [[HLA-DR3]] | ||
| | | | ||
* Associated with an overall 2 to 3 fold increase in the risk of SLE | * Associated with an overall 2- to 3-fold increase in the risk of SLE | ||
* More in European and Asian people | * More common in European and Asian people | ||
* [[HLA-DQ]] and [[HLA-DR]] alleles: | * [[HLA-DQ]] and [[HLA-DR]] alleles: | ||
** Strong association with SLE [[autoantibodies]] | ** Strong association with SLE [[autoantibodies]] | ||
|- | |- | ||
! rowspan="2" style="background:#DCDCDC;" |Immune complex dependent response | ! rowspan="2" style="background:#DCDCDC;" |Immune complex dependent response | ||
| | | style="background:#DCDCDC;" align="center" |[[HLA]] class 3<ref name="pmid11079100">{{cite journal |vauthors=Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ |title=Systemic lupus erythematosus, complement deficiency, and apoptosis |journal=Adv. Immunol. |volume=76 |issue= |pages=227–324 |year=2000 |pmid=11079100 |doi= |url=}}</ref> | ||
| | | | ||
* Contains important [[Gene|immune genes]] including: | * Contains important [[Gene|immune genes]] including: | ||
| Line 150: | Line 146: | ||
* Circulating complement C4 proteins deficiency will promote [[autoimmunity]] | * Circulating complement C4 proteins deficiency will promote [[autoimmunity]] | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |C1q genes<ref name="pmid11079100">{{cite journal |vauthors=Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ |title=Systemic lupus erythematosus, complement deficiency, and apoptosis |journal=Adv. Immunol. |volume=76 |issue= |pages=227–324 |year=2000 |pmid=11079100 |doi= |url=}}</ref> | ||
| colspan="1" rowspan="1" | | | colspan="1" rowspan="1" | | ||
* The [[complement system]] acts through [[opsonization]]: | * The [[complement system]] acts through [[opsonization]]: | ||
| Line 162: | Line 158: | ||
|- | |- | ||
! rowspan="4" style="background:#DCDCDC;" |Innate response | ! rowspan="4" style="background:#DCDCDC;" |Innate response | ||
| | | style="background:#DCDCDC;" align="center" |[[IRF5|Interferon (IFN) regulatory factor 5]]<ref name="pmid20080916">{{cite journal |vauthors=Löfgren SE, Yin H, Delgado-Vega AM, Sanchez E, Lewén S, Pons-Estel BA, Witte T, D'Alfonso S, Ortego-Centeno N, Martin J, Alarcón-Riquelme ME, Kozyrev SV |title=Promoter insertion/deletion in the IRF5 gene is highly associated with susceptibility to systemic lupus erythematosus in distinct populations, but exerts a modest effect on gene expression in peripheral blood mononuclear cells |journal=J. Rheumatol. |volume=37 |issue=3 |pages=574–8 |year=2010 |pmid=20080916 |doi=10.3899/jrheum.090440 |url=}}</ref> | ||
| colspan="1" rowspan="1" | | | colspan="1" rowspan="1" | | ||
* Codes a [[transcription factor]] in the type 1 [[interferon]] pathway | * Codes a [[transcription factor]] in the type 1 [[interferon]] pathway | ||
| Line 174: | Line 170: | ||
* Specific combinations of several [[polymorphisms]] in the [[IRF5]] region interact to increase disease risk | * Specific combinations of several [[polymorphisms]] in the [[IRF5]] region interact to increase disease risk | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[STAT4]]<ref name="pmid18579578">{{cite journal |vauthors=Sigurdsson S, Nordmark G, Garnier S, Grundberg E, Kwan T, Nilsson O, Eloranta ML, Gunnarsson I, Svenungsson E, Sturfelt G, Bengtsson AA, Jönsen A, Truedsson L, Rantapää-Dahlqvist S, Eriksson C, Alm G, Göring HH, Pastinen T, Syvänen AC, Rönnblom L |title=A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5 |journal=Hum. Mol. Genet. |volume=17 |issue=18 |pages=2868–76 |year=2008 |pmid=18579578 |pmc=2525501 |doi=10.1093/hmg/ddn184 |url=}}</ref><ref name="pmid19109131">{{cite journal |vauthors=Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB |title=Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo |journal=J. Immunol. |volume=182 |issue=1 |pages=34–8 |year=2009 |pmid=19109131 |pmc=2716754 |doi= |url=}}</ref><ref name="pmid18516230">{{cite journal |vauthors=Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, Chung SA, Nititham J, Hom G, Kao AH, Demirci FY, Kamboh MI, Petri M, Manzi S, Kastner DL, Seldin MF, Gregersen PK, Behrens TW, Criswell LA |title=Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus |journal=PLoS Genet. |volume=4 |issue=5 |pages=e1000084 |year=2008 |pmid=18516230 |pmc=2377340 |doi=10.1371/journal.pgen.1000084 |url=}}</ref><ref name="pmid18803832">{{cite journal |vauthors=Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, Matsumoto I, Ito S, Tsutsumi A, Koga M, Arinami T, Graham RR, Hom G, Takasaki Y, Hashimoto H, Behrens TW, Sumida T, Tsuchiya N |title=Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region |journal=Arthritis Res. Ther. |volume=10 |issue=5 |pages=R113 |year=2008 |pmid=18803832 |pmc=2592800 |doi=10.1186/ar2516 |url=}}</ref> | ||
| | | | ||
* Encodes the signal transducer and activator of transcription 4 protein | * Encodes the signal transducer and activator of transcription 4 protein | ||
| Line 184: | Line 180: | ||
* [[Mutation]] lead to an increased [[sensitivity]] to [[Interferon-alpha|IFN-α]] signaling in peripheral blood [[mononuclear cells]] | * [[Mutation]] lead to an increased [[sensitivity]] to [[Interferon-alpha|IFN-α]] signaling in peripheral blood [[mononuclear cells]] | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |The ''[[IRAK1]]-[[MECP2]]'' region | ||
| | | | ||
* | * Encodes a [[protein kinase]]: | ||
** Regulates multiple pathways in both [[Innate immune system|innate]] and [[Adaptive immune response|adaptive immune responses]] by linking several immune-receptor-complexes to [[TNF]] receptor-associated factor 6 | ** Regulates multiple pathways in both [[Innate immune system|innate]] and [[Adaptive immune response|adaptive immune responses]] by linking several immune-receptor-complexes to [[TNF]] receptor-associated factor 6 | ||
** Critical role in the [[Transcriptional regulation|transcriptional suppression]] of [[methylation]]-sensitive genes | ** Critical role in the [[Transcriptional regulation|transcriptional suppression]] of [[methylation]]-sensitive genes | ||
| Line 192: | Line 188: | ||
* The exact [[Pathogenicity|pathogenetics]] is not completely known | * The exact [[Pathogenicity|pathogenetics]] is not completely known | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[Fc region|FcγR genes]]<ref name="pmid10413210">{{cite journal |vauthors=Yap SN, Phipps ME, Manivasagar M, Tan SY, Bosco JJ |title=Human Fc gamma receptor IIA (FcgammaRIIA) genotyping and association with systemic lupus erythematosus (SLE) in Chinese and Malays in Malaysia |journal=Lupus |volume=8 |issue=4 |pages=305–10 |year=1999 |pmid=10413210 |doi=10.1191/096120399678847876 |url=}}</ref> | ||
| | | | ||
* Encode [[proteins]] that: | * Encode [[proteins]] that: | ||
** Recognize [[immune complexes]] | ** Recognize [[immune complexes]] | ||
** | ** Are involved in [[Antibody-dependent cellular cytotoxicity|antibody-dependent]] responses | ||
| | | | ||
* [[Mutation]] associated with: | * [[Mutation]] associated with: | ||
| Line 203: | Line 199: | ||
|- | |- | ||
! rowspan="2" style="background:#DCDCDC;" |Cell apoptosis regulators | ! rowspan="2" style="background:#DCDCDC;" |Cell apoptosis regulators | ||
| | | style="background:#DCDCDC;" align="center" |[[TREX1]] | ||
| | | | ||
* Encodes a major [[exonuclease]]: | * Encodes a major [[exonuclease]]: | ||
| Line 216: | Line 212: | ||
*# Systemic [[autoimmunity]] | *# Systemic [[autoimmunity]] | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[Interleukin 10|IL-10]] | ||
| | | | ||
* Encodes [[IL-10]] | * Encodes [[IL-10]] | ||
| Line 224: | Line 220: | ||
|- | |- | ||
! rowspan="2" style="background:#DCDCDC;" |IFNα regulators | ! rowspan="2" style="background:#DCDCDC;" |IFNα regulators | ||
| | | style="background:#DCDCDC;" align="center" |[[TNFAIP3]] and [[TNIP1]] | ||
| | | | ||
* | * Encode key regulators of the [[NFκB]] signaling pathway | ||
* Modulate cell activation, [[cytokine]] signaling and [[apoptosis]] | * Modulate cell activation, [[cytokine]] signaling and [[apoptosis]] | ||
| | | | ||
* The exact [[Pathogenicity|pathogenetics]] is not completely known | * The exact [[Pathogenicity|pathogenetics]] is not completely known | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |PHRF1 | ||
| | | | ||
* Encodes an [[elongation factor]] | * Encodes an [[elongation factor]] | ||
| Line 238: | Line 234: | ||
|- | |- | ||
! rowspan="7" style="background:#DCDCDC;" |Regulators of Lymphocytes | ! rowspan="7" style="background:#DCDCDC;" |Regulators of Lymphocytes | ||
| | | style="background:#DCDCDC;" align="center" |TNFSF4 | ||
| | | | ||
* The [[genes]] in this loci produce interaction induces the production of co-stimulatory signals to activate [[T cells]] | * The [[genes]] in this loci produce interaction induces the production of co-stimulatory signals to activate [[T cells]] | ||
| Line 249: | Line 245: | ||
** Influencing the functional consequences of [[T cell|T-cell activation]] | ** Influencing the functional consequences of [[T cell|T-cell activation]] | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[BLK (gene)|BLK]]<ref name="pmid19180478">{{cite journal |vauthors=Ito I, Kawasaki A, Ito S, Hayashi T, Goto D, Matsumoto I, Tsutsumi A, Hom G, Graham RR, Takasaki Y, Hashimoto H, Ohashi J, Behrens TW, Sumida T, Tsuchiya N |title=Replication of the association between the C8orf13-BLK region and systemic lupus erythematosus in a Japanese population |journal=Arthritis Rheum. |volume=60 |issue=2 |pages=553–8 |year=2009 |pmid=19180478 |doi=10.1002/art.24246 |url=}}</ref> | ||
| | | | ||
* Encodes a [[protein kinase]]: | * Encodes a [[protein kinase]]: | ||
** Mediates [[intracellular signaling]] | ** Mediates [[intracellular signaling]] | ||
** Influences [[B cells]] proliferation and differentiation | ** Influences [[B cells]] proliferation and differentiation | ||
** | ** Influences tolerance of [[B cells]] | ||
| | | | ||
* More common in Chinese and Japanese populations | * More common in Chinese and Japanese populations | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[PTPN22]]<ref name="pmid19302045">{{cite journal |vauthors=Gregersen PK, Olsson LM |title=Recent advances in the genetics of autoimmune disease |journal=Annu. Rev. Immunol. |volume=27 |issue= |pages=363–91 |year=2009 |pmid=19302045 |pmc=2992886 |doi=10.1146/annurev.immunol.021908.132653 |url=}}</ref> | ||
| | | | ||
* Encodes a [[lymphoid]]-specific [[Phosphatases|phosphatase]] that inhibits [[T cell|T-cell]] activation | * Encodes a [[lymphoid]]-specific [[Phosphatases|phosphatase]] that inhibits [[T cell|T-cell]] activation | ||
| Line 265: | Line 261: | ||
* More seen in European populations | * More seen in European populations | ||
* [[Mutation]] increases the intrinsic [[lymphoid]]-specific [[Phosphatases|phosphatase]] activity that lead to: | * [[Mutation]] increases the intrinsic [[lymphoid]]-specific [[Phosphatases|phosphatase]] activity that lead to: | ||
** | ** Reduction of [[T cell receptor|T-cell receptor (TCR)]] signaling threshold | ||
** | ** Promotion of [[autoimmunity]] | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |BANK1<ref name="pmid11782428">{{cite journal |vauthors=Yokoyama K, Su Ih IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, Yamamoto T |title=BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor |journal=EMBO J. |volume=21 |issue=1-2 |pages=83–92 |year=2002 |pmid=11782428 |pmc=125810 |doi=10.1093/emboj/21.1.83 |url=}}</ref><ref name="pmid18204447">{{cite journal |vauthors=Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barizzone N, Barrizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, González-Escribano MF, Martin J, Abderrahim H, Alarcón-Riquelme ME |title=Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus |journal=Nat. Genet. |volume=40 |issue=2 |pages=211–6 |year=2008 |pmid=18204447 |doi=10.1038/ng.79 |url=}}</ref> | ||
| | | | ||
* Encodes a [[B cell|B-cell]] adaptor protein | * Encodes a [[B cell|B-cell]] adaptor protein | ||
* Facilitates the release of [[intracellular]] [[calcium]] | * Facilitates the release of [[intracellular]] [[calcium]] | ||
* | * Alters the [[B-cell]] activation threshold | ||
| rowspan="2" | | | rowspan="2" | | ||
* Mutations lead to hyperctivation of [[B-cell receptor|B-cell receptors]] and the subsequent [[B-cell]] hyperactivity that is commonly observed in SLE | * Mutations lead to hyperctivation of [[B-cell receptor|B-cell receptors]] and the subsequent [[B-cell]] hyperactivity that is commonly observed in SLE | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |LYN<ref name="pmid18204446">{{cite journal |vauthors=Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA |title=Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci |journal=Nat. Genet. |volume=40 |issue=2 |pages=204–10 |year=2008 |pmid=18204446 |pmc=3712260 |doi=10.1038/ng.81 |url=}}</ref> | ||
| | | | ||
* Mediates [[B-cell]] activation | * Mediates [[B-cell]] activation | ||
* Mediates [[B-cell]] inhibition | * Mediates [[B-cell]] inhibition | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |ETS1<ref name="pmid17967903">{{cite journal |vauthors=Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC |title=Ets-1 is a negative regulator of Th17 differentiation |journal=J. Exp. Med. |volume=204 |issue=12 |pages=2825–35 |year=2007 |pmid=17967903 |pmc=2118518 |doi=10.1084/jem.20070994 |url=}}</ref><ref name="pmid19838195">{{cite journal |vauthors=Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jönsen A, Bengtsson AA, Rantapää-Dahlqvist S, Baechler EC, Brown EE, Alarcón GS, Edberg JC, Ramsey-Goldman R, McGwin G, Reveille JD, Vilá LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Rönnblom L, Criswell LA, Syvänen AC, Behrens TW, Graham RR |title=A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus |journal=Nat. Genet. |volume=41 |issue=11 |pages=1228–33 |year=2009 |pmid=19838195 |pmc=2925843 |doi=10.1038/ng.468 |url=}}</ref> | ||
| | | | ||
* Negatively regulates the differentiation of [[B cells]] and type 17 [[T-helper cells]] | * Negatively regulates the differentiation of [[B cells]] and type 17 [[T-helper cells]] | ||
| Line 288: | Line 284: | ||
* The exact [[Pathogenicity|pathogenetics]] is not completely known | * The exact [[Pathogenicity|pathogenetics]] is not completely known | ||
|- | |- | ||
| | | style="background:#DCDCDC;" align="center" |[[IKZF1]]<ref name="pmid17357110">{{cite journal |vauthors=Wojcik H, Griffiths E, Staggs S, Hagman J, Winandy S |title=Expression of a non-DNA-binding Ikaros isoform exclusively in B cells leads to autoimmunity but not leukemogenesis |journal=Eur. J. Immunol. |volume=37 |issue=4 |pages=1022–32 |year=2007 |pmid=17357110 |doi=10.1002/eji.200637026 |url=}}</ref> | ||
| | | | ||
* | * Encodes a [[lymphoid]]-restricted [[transcription factor]] | ||
* Regulates: | * Regulates: | ||
** [[Lymphocyte]] differentiation and proliferation | ** [[Lymphocyte]] differentiation and proliferation | ||
| Line 299: | Line 295: | ||
|- | |- | ||
! style="background:#DCDCDC;" |Genes involved in immune complex clearance | ! style="background:#DCDCDC;" |Genes involved in immune complex clearance | ||
| | | style="background:#DCDCDC;" align="center" |ITGAM<ref name="pmid19838195">{{cite journal |vauthors=Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jönsen A, Bengtsson AA, Rantapää-Dahlqvist S, Baechler EC, Brown EE, Alarcón GS, Edberg JC, Ramsey-Goldman R, McGwin G, Reveille JD, Vilá LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Rönnblom L, Criswell LA, Syvänen AC, Behrens TW, Graham RR |title=A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus |journal=Nat. Genet. |volume=41 |issue=11 |pages=1228–33 |year=2009 |pmid=19838195 |pmc=2925843 |doi=10.1038/ng.468 |url=}}</ref> | ||
| | | | ||
* | * Encodes a [[protein]] that binds the [[complement]] cleavage fragment of [[C3b]] | ||
| | | | ||
* | * Contributes to SLE susceptibility | ||
|} | |} | ||
| Line 313: | Line 309: | ||
* Anti-Ro, anti-La, anti sm, and anti RNP [[antibodies]] have been associated with [[mucocutaneous]] involvement and less severe [[nephropathy]]. | * Anti-Ro, anti-La, anti sm, and anti RNP [[antibodies]] have been associated with [[mucocutaneous]] involvement and less severe [[nephropathy]]. | ||
== Gross Pathology == | == Gross Pathology == | ||
On gross pathology | On gross pathology the most important characteristic findings are: | ||
* [[Kidney]]: Bilateral [[pallor]] and [[Hypertrophy (medical)|hypertrophy]] | * [[Kidney]]: Bilateral [[pallor]] and [[Hypertrophy (medical)|hypertrophy]] | ||
* [[Brain]]: [[Infarct]] regions and [[hemorrhages]] | * [[Brain]]: [[Infarct]] regions and [[hemorrhages]] | ||
* [[ | * [[Heart]]: [[Cardiomegaly]] and [[valvular]] vegetation | ||
* [[Pleura]]: [[Pleural fibrosis| | * [[Pleura|Lung]]: [[Pleural fibrosis|Peural fibrosis]] and [[pleural effusion]] | ||
== Microscopic Pathology == | == Microscopic Pathology == | ||
On microscopic histopathological analysis, lupus erythematosus (LE) cells can be seen in SLE. LE cells are [[neutrophils]] that have engulfed an intact nucleus. LE cells are also known as LE bodies. | |||
On microscopic [[histopathological]] analysis, [[apoptotic]] [[keratinocytes]], [[vacuolization]] of the [[basement membrane]], and dermal mucin deposition are characteristic findings of SLE [[dermatitis]], and active or inactive [[Endocapillary proliferative glomerulonephritis|endocapillary]] or extracapillary segmental [[glomerulonephritis]] are characteristic findings of [[Lupus nephritis|SLE nephritis]]. [[Microscopic]] findings in systemic lupus erythematosus based on organ system | On microscopic [[histopathological]] analysis, [[apoptotic]] [[keratinocytes]], [[vacuolization]] of the [[basement membrane]], and dermal mucin deposition are characteristic findings of SLE [[dermatitis]], and active or inactive [[Endocapillary proliferative glomerulonephritis|endocapillary]] or extracapillary segmental [[glomerulonephritis]] are characteristic findings of [[Lupus nephritis|SLE nephritis]]. [[Microscopic]] findings in systemic lupus erythematosus are based on the involved organ system. | ||
=== Skin | === Skin histopathology === | ||
Common shared [[histopathologic]] features among all different subtypes of cutaneous lupus include: | Common shared [[histopathologic]] features among all different subtypes of cutaneous lupus include: | ||
* [[Hyperkeratosis]] | * [[Hyperkeratosis]] | ||
| Line 335: | Line 331: | ||
* [[Superficial]], [[Perivascular cell|perivascular]], and perifollicular areas (due to [[Mononuclear cells|mononuclear cell]] inflammatory infiltrate) | * [[Superficial]], [[Perivascular cell|perivascular]], and perifollicular areas (due to [[Mononuclear cells|mononuclear cell]] inflammatory infiltrate) | ||
{| class="wikitable" | {| class="wikitable" | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |SLE dermatitis subtype | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Specific microscopic findings | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Preview | ||
|- | |- | ||
! | ! style="background:#DCDCDC;" align="center" |Acute cutaneous lupus erythematosus | ||
| | | | ||
* Lymphohistiocytic infiltrate in the [[superficial]] dermis <ref name="pmid20482683">{{cite journal |vauthors=Sepehr A, Wenson S, Tahan SR |title=Histopathologic manifestations of systemic diseases: the example of cutaneous lupus erythematosus |journal=J. Cutan. Pathol. |volume=37 Suppl 1 |issue= |pages=112–24 |year=2010 |pmid=20482683 |doi=10.1111/j.1600-0560.2010.01510.x |url=}}</ref> | * Lymphohistiocytic infiltrate in the [[superficial]] dermis <ref name="pmid20482683">{{cite journal |vauthors=Sepehr A, Wenson S, Tahan SR |title=Histopathologic manifestations of systemic diseases: the example of cutaneous lupus erythematosus |journal=J. Cutan. Pathol. |volume=37 Suppl 1 |issue= |pages=112–24 |year=2010 |pmid=20482683 |doi=10.1111/j.1600-0560.2010.01510.x |url=}}</ref> | ||
| rowspan="3" |[[File:Vacuolar interface dermatitis - high mag.jpg|thumb|300px|<SMALL><SMALL>''[https://librepathology.org/wiki/Main_Page/ Adapted from Librepathology]''</SMALL></SMALL>]] | | rowspan="3" |[[File:Vacuolar interface dermatitis - high mag.jpg|thumb|300px|<SMALL><SMALL>''[https://librepathology.org/wiki/Main_Page/ Adapted from Librepathology]''</SMALL></SMALL>]] | ||
|- | |- | ||
! | ! style="background:#DCDCDC;" align="center" |Subacute cutaneous lupus erythematosus | ||
| | | | ||
* Less [[Follicular cell|follicular]] plugging and [[hyperkeratosis]] in comparison with dischoid lupus erythematosus | * Less [[Follicular cell|follicular]] plugging and [[hyperkeratosis]] in comparison with dischoid lupus erythematosus | ||
| Line 350: | Line 346: | ||
* Absence or minimal change of [[basement membrane]] thickening | * Absence or minimal change of [[basement membrane]] thickening | ||
|- | |- | ||
! | ! style="background:#DCDCDC;" align="center" |Chronic cutaneous lupus erythematosus | ||
| | | | ||
* [[Discoid lupus erythematosus]] : | * [[Discoid lupus erythematosus]] : | ||
| Line 366: | Line 362: | ||
=== Glomerulonephritis histopathology === | === Glomerulonephritis histopathology === | ||
{| class="wikitable" | {| class="wikitable" | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |<small>Class<small> | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |SLE nephritis subtype | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Light microscopy findings | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Light microscopy previews | ||
! | ! style="background: #4479BA; color: #FFFFFF; " align="center" |Electron microscopy/Immunofluorescence findings | ||
|- | |- | ||
! align="center" |'''I''' | ! align="center" |'''I''' | ||
| Line 429: | Line 425: | ||
|} | |} | ||
=== Synovial | === Synovial histopathology === | ||
* Nonspecific histopathologic findings | * Nonspecific histopathologic findings | ||
* Superficial [[fibrin]]-like material | * Superficial [[fibrin]]-like material | ||
| Line 439: | Line 435: | ||
** [[Thrombi]] | ** [[Thrombi]] | ||
=== Mucosal | === Mucosal histopathology === | ||
* [[Hyperkeratosis]] | * [[Hyperkeratosis]] | ||
* [[Atrophy]] of rete processes | * [[Atrophy]] of rete processes | ||
| Line 450: | Line 446: | ||
==== | =====Lupus nephritis histopathology===== | ||
{{#ev:youtube|Tw07BFaDEo0}} | {{#ev:youtube|Tw07BFaDEo0}} | ||
Latest revision as of 18:29, 23 August 2017
| Title |
| https://https://www.youtube.com/watch?v=0junqD4BLH4%7C350}} |
|
Systemic lupus erythematosus Microchapters |
|
Differentiating Systemic lupus erythematosus from other Diseases |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Systemic lupus erythematosus pathophysiology On the Web |
|
American Roentgen Ray Society Images of Systemic lupus erythematosus pathophysiology |
|
Directions to Hospitals Treating Systemic lupus erythematosus |
|
Risk calculators and risk factors for Systemic lupus erythematosus pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Mahshid Mir, M.D. [2], Cafer Zorkun, M.D., Ph.D. [3], Raviteja Guddeti, M.B.B.S. [4]
Overview
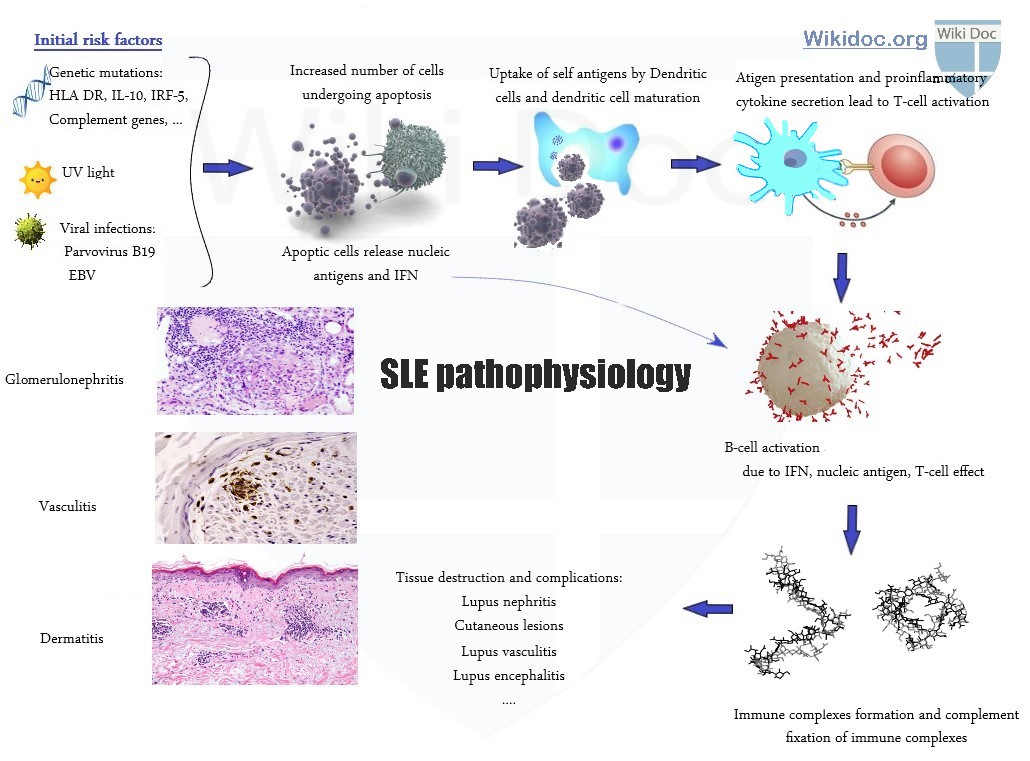
The pathophysiology of systemic lupus erythematosus involves the immune system. Other factors such as genetic factors, hormonal abnormalities, and environmental factors also play a role. The most important environmental factors involved in the pathogenesis of SLE include ultraviolet (UV) light and some infections. The most important genes involved in the pathogenesis of SLE include HLA-DR2, HLA-DR3, HLA class 3, C1q, and interferon (IFN) regulatory factor 5. The most prominent events involving immune abnormalities are related to persistent activation of B cells and plasma cells that make auto-antibodies during disease progression. The disease developmental process begins with the release of microparticles and proinflammatory cytokines from the cells that are undergoing apoptosis. Due to excess amount of apoptosis, the body is unable to clear these microparticles entirely, and these microparticles are presented to dendritic cells as antigens. Dendritic cells process these microparticles and mature, and present these as antigens to T-cells. T-cells, microparticles, and proinflammatory cytokines themselves trigger B-cell activation and autoantibody production. As a result, body tissues lose their self-tolerance. The most prominent events involving hormonal abnormalities are due to prolactin and estrogen. On microscopic histopathological analysis, apoptotic keratinocytes, vacuolization of the basement membrane, and dermal mucin deposition are characteristic findings of SLE dermatitis, and active or inactive endocapillary or extracapillary segmental glomerulonephritis are characteristic findings of lupus nephritis.
Pathogenesis
The progression of systemic lupus erythematosus (SLE) involves the immune system. Nearly all of the pathological manifestations of SLE occur due to antibody formation and the creation and deposition of immune complexes in different organs of the body. When the immune complexes are formed, they deposit on different body tissues and vessels, which may lead to complement activation and more organ damage. There are other factors such as genetic factors, hormonal abnormalities, and environmental factors that also play a role in the pathogenesis of SLE.
Environmental factors
The environmental factors and genetic factors are the most important risk factors for developing SLE because they may jump-start the disinhibited cellular apoptosis chain. This apoptosis step is the first step in the pathogenesis of lupus.
- Infections
- May stimulate some antigen specific cells and increase apoptosis
- May induce anti-DNA antibodies
- May mimic lupus-like symptoms
- Associated with higher risk of SLE
- Associated with triggering the active courses and flare ups of disease in children
- Include:
- Parvovirus B19
- Epstein-Barr virus (EBV)
- Trypanosomiasis
- Mycobacterial infections
- SLE flares may follow bacterial infections
- Ultraviolet (UV) light:
- Can stimulate B-cells to produce more antibodies
- May activate macrophages, interfere with antigen processing, and therefore increase the degree of autoimmunity
Immune abnormalities
The development of systemic lupus erythematosus (SLE) is due to the activation of different mechanisms that may result in auto-immunity. The disease developmental process begins with the release of microparticles and proinflammatory cytokines from the cells that are undergoing apoptosis. Due to excess amount of apoptosis, the body is unable to clear these microparticles entirely, and these microparticles are presented to dendritic cells as antigens. Dendritic cells process these microparticles and mature, and present these as antigens to T-cells. T-cells, microparticles, and proinflammatory cytokines themselves trigger B-cell activation and autoantibody production. As a result, body tissues lose their self-tolerance. Affected patients are no longer entirely tolerant to all of their self-antigens, leading to development of an autoimmune disease and producing autoantibodies as a response. During disease progression, B cells and plasma cells that make autoantibodies are more persistently activated due to signaling abnormalities, causing them to make more autoantibodies. These autoantibodies are targeted predominantly to intracellular nucleoprotein particles.[1][2] This increase in autoantibody production and persistence is supposed to be downregulated by anti-idiotypic antibodies or regulatory immune cells, but the massive immunologic response in SLE prevents this downregulation from taking place. After formation of immune complexes, the classical complement pathway is activated, which leads to the deposition of immune complexes in different organs and is responsible for flare ups and long term complications. The most important immune abnormalities that are related to SLE development and progression are:
Microparticles
Increased level of microparticles (MPs):[3]
- Microparticles are small, membrane-bound vesicles enclosing DNA, RNA, nuclear proteins, cell adhesion molecules, growth factors, and cytokines
- They are shed from cells during apoptosis or activation
- Microparticles can drive inflammation and autoimmunity by their derivatives
Pro-inflammatory cytokines
Increased expression of specific genetic factors may be associated with promoting autoimmunity. The most important cytokine changes include:[4][3]
- Increased expression of interferon alpha (IFN-α) inducible RNA transcripts by mononuclear cells
- Increased IFN-I production due to increased availability of stimulatory nucleic acids
- May be responsible for SLE chronic characteristics
- Elevated levels of circulating TNF-alpha (expressed by renal tissue in lupus nephritis) correlate with active disease
Signaling abnormalities
Protein kinases are responsible for intracellular cytokine signals. Intracellular signaling leads to various types of cell response, such as:
Cell signaling abnormalities leads to:
- T and B lymphocytes cellular hyperactivity
- T and B lymphocytes hyper responsiveness
- Persistence of auto-reactive T cells that would otherwise have been deleted
Signaling abnormalities of T and B lymphocytes, may be due to:
- Abnormal voltage-gated potassium channels, these channels facilitate excessive calcium entry into T cells and lead to increased calcium responses to antigen stimulation
- Hyperphosphorylation of cytosolic protein substrates
- Decreased nuclear factor kB
B-Cell role
- Increase in circulating plasma cells and memory B cells that are associated with SLE activity
- Polyclonal activation of B cells and abnormal B-cell receptor signaling
- Increase in B cells' life span
T-Cell role
- Decrease in cytotoxic T cells, decrease in suppressor T cell function, and impaired generation of polyclonal T-cell cytolytic activity
- Increased number and activity of helper T cells
Neutrophil role
- Increased number of circulating neutrophils undergoing NETosis (NET=neutrophil extracellular traps), a form of apoptosis specific for neutrophils, releases DNA bound to protein in protein nets, which stimulates anti-DNA and IFN-alpha production
- Increased neutrophil extracellular trap leads to: [5]
- Thrombus formation
- Increased disease activity and renal disease and thus can be used even as a disease activity marker
- Endothelial cell damage and inflammation in atherosclerotic plaques, which may contribute to accelerated atherosclerosis in systemic lupus erythematosus
Hormonal abnormalities
The following evidence is suggestive of the hormonal predisposition to SLE:
- Predilection of the disease for females shows the relationship between female hormones and the onset of SLE
- Significantly increased risk for SLE in:[6]
- Early age of menarche
- Early age at menopause or surgical menopause
- Women that are treated with estrogen-containing regimens such as oral contraceptives or postmenopausal hormone replacement therapies
Hormones that are related to disease progression include:[7]
Prolactin:
- Stimulates the immune system and is elevated in SLE
Exogenous estrogen
- Including oral contraceptive use and postmenopausal hormone replacement therapy: [7][8]
- Stimulates the type 1 IFN pathway
- Stimulates thymocytes, CD8+ and CD4+ T cells, B cells, macrophages, and causes the release of certain cytokines (eg, IL-1)
- Prompt maturation of B cells, especially those that have a high affinity to anti-DNA antibodies by decreasing the apoptosis of self-reactive B-cells[9]
- Stimulate expression of HLA and endothelial cell adhesion molecules (VCAM, ICAM)
- Increases macrophage proto-oncogene expression
- Enhanced adhesion of peripheral mononuclear cells to endothelium
Progesterone:
- May inhibit the type 1 interferon pathway, suggesting that a balance between estrogen and progesterone may be critical for the body to remain healthy
- Both progesterone and high levels of estrogen promote a Th2 response, which favors auto-antibody production
Genetics
Systemic lupus erythematosus is transmitted in a polygenic inheritance pattern. Genes involved in the pathogenesis of systemic lupus erythematosus include HLA class 2 (especially DR2 and DR3), HLA class 3 (especially complement genes including C2 and C4 genes), IFNRF5 gene, and other genes related to the immunologic system. The following evidence is also suggestive of the genetic predisposition of SLE:[10]
- Increase occurrence of disease in identical twins
- Increased disease frequency among first degree relatives
- The increased occurrence of the disease in siblings of SLE patients
| Class | Gene subtype | Function | Pathological effect and Molecular mechanisms |
|---|---|---|---|
| Autoantigen presentation | HLA class 2[11] |
|
|
| Immune complex dependent response | HLA class 3[12] |
|
|
| C1q genes[12] |
|
| |
| Innate response | Interferon (IFN) regulatory factor 5[13] |
|
|
| STAT4[14][15][16][17] |
|
| |
| The IRAK1-MECP2 region |
|
| |
| FcγR genes[18] |
|
| |
| Cell apoptosis regulators | TREX1 |
|
|
| IL-10 |
|
||
| IFNα regulators | TNFAIP3 and TNIP1 |
| |
| PHRF1 |
|
| |
| Regulators of Lymphocytes | TNFSF4 |
| |
| BLK[19] |
|
| |
| PTPN22[20] |
|
| |
| BANK1[21][22] |
|
| |
| LYN[23] | |||
| ETS1[24][25] |
|
| |
| IKZF1[26] |
|
| |
| Genes involved in immune complex clearance | ITGAM[25] |
|
|
Associated Conditions
- Homozygous deficiencies of the components of complement, especially C1q, are associated with developing immunologic diseases, particularly SLE or a lupus-like disease.[27]
- The FcγRIIA polymorphism has been associated with nephritis in African Americans, Koreans, and Hispanics. Both FcgammaRIIa and FcgammaRIIIa have low binding alleles that confer risk for SLE and may act in the pathogenesis of disease. [28]
- Women treated with estrogen-containing regimens such as oral contraceptives or postmenopausal hormone replacement therapies are more predisposed to SLE.
- Annular or psoriasiform skin lesions are associated with anti-Ro (SS-A) and anti-La (SS-B) antibodies.
- Anti-Ro, anti-La, anti sm, and anti RNP antibodies have been associated with mucocutaneous involvement and less severe nephropathy.
Gross Pathology
On gross pathology the most important characteristic findings are:
- Kidney: Bilateral pallor and hypertrophy
- Brain: Infarct regions and hemorrhages
- Heart: Cardiomegaly and valvular vegetation
- Lung: Peural fibrosis and pleural effusion
Microscopic Pathology
On microscopic histopathological analysis, lupus erythematosus (LE) cells can be seen in SLE. LE cells are neutrophils that have engulfed an intact nucleus. LE cells are also known as LE bodies.
On microscopic histopathological analysis, apoptotic keratinocytes, vacuolization of the basement membrane, and dermal mucin deposition are characteristic findings of SLE dermatitis, and active or inactive endocapillary or extracapillary segmental glomerulonephritis are characteristic findings of SLE nephritis. Microscopic findings in systemic lupus erythematosus are based on the involved organ system.
Skin histopathology
Common shared histopathologic features among all different subtypes of cutaneous lupus include:
- Hyperkeratosis
- Epidermal atrophy
- Dermal mucin deposition
- Liquefactive degeneration of the basal layer of the epidermis and vacuolization
- Thickening of the basement membrane
- Pigment incontinence
- Mononuclear cell infiltration at dermo-epidermal junction
- Superficial, perivascular, and perifollicular areas (due to mononuclear cell inflammatory infiltrate)
| SLE dermatitis subtype | Specific microscopic findings | Preview |
|---|---|---|
| Acute cutaneous lupus erythematosus |
|
 |
| Subacute cutaneous lupus erythematosus |
| |
| Chronic cutaneous lupus erythematosus |
|
Glomerulonephritis histopathology
| Class | SLE nephritis subtype | Light microscopy findings | Light microscopy previews | Electron microscopy/Immunofluorescence findings |
|---|---|---|---|---|
| I | Minimal mesangial lupus nephritis | - |
| |
| II | Mesangial proliferative lupus nephritis |
|
 |
|
| III | Focal lupus nephritis |
|
 |
|
| IV | Diffuse lupus nephritis |
|
 |
|
| V | Lupus membranous nephropathy |
|
| |
| VI | Advanced sclerosing lupus nephritis |  |
|
Synovial histopathology
- Nonspecific histopathologic findings
- Superficial fibrin-like material
- Local or diffuse synovial cell lining proliferation
- Vascular changes:
- Perivascular mononuclear cells
- Lumen obliteration
- Enlarged endothelial cells
- Thrombi
Mucosal histopathology
- Hyperkeratosis
- Atrophy of rete processes
- Superficial and deep inflammatory infiltrates
- Edema in the lamina propria
- Continuous or patchy periodic acid-Schiff (PAS)-positive deposits in the basement membrane zone
- Deposition of intercellular mucin
- Deposition of immunoglobulin and complement at the dermal-epidermal junction
Lupus nephritis histopathology
{{#ev:youtube|Tw07BFaDEo0}}
References
- ↑ Elkon K (1995). "Autoantibodies in systemic lupus erythematosus". Curr Opin Rheumatol. 7 (5): 384–8. PMID 8519610.
- ↑ Yaniv G, Twig G, Shor DB, Furer A, Sherer Y, Mozes O, Komisar O, Slonimsky E, Klang E, Lotan E, Welt M, Marai I, Shina A, Amital H, Shoenfeld Y (2015). "A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients". Autoimmun Rev. 14 (1): 75–9. doi:10.1016/j.autrev.2014.10.003. PMID 25449682.
- ↑ 3.0 3.1 Dye JR, Ullal AJ, Pisetsky DS (2013). "The role of microparticles in the pathogenesis of rheumatoid arthritis and systemic lupus erythematosus". Scand. J. Immunol. 78 (2): 140–8. doi:10.1111/sji.12068. PMID 23672591.
- ↑ Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, Ly N, Woodward RN, Fry KE, Lau AY, Prentice JG, Wohlgemuth JG, Crow MK (2004). "Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus". Arthritis Rheum. 50 (12): 3958–67. doi:10.1002/art.20798. PMID 15593221.
- ↑ Barnado A, Crofford LJ, Oates JC (2016). "At the Bedside: Neutrophil extracellular traps (NETs) as targets for biomarkers and therapies in autoimmune diseases". J. Leukoc. Biol. 99 (2): 265–78. doi:10.1189/jlb.5BT0615-234R. PMID 26658004.
- ↑ Costenbader KH, Feskanich D, Stampfer MJ, Karlson EW (2007). "Reproductive and menopausal factors and risk of systemic lupus erythematosus in women". Arthritis Rheum. 56 (4): 1251–62. doi:10.1002/art.22510. PMID 17393454.
- ↑ 7.0 7.1 Lahita RG (1999). "The role of sex hormones in systemic lupus erythematosus". Curr Opin Rheumatol. 11 (5): 352–6. PMID 10503654.
- ↑ Hughes GC, Choubey D (2014). "Modulation of autoimmune rheumatic diseases by oestrogen and progesterone". Nat Rev Rheumatol. 10 (12): 740–51. doi:10.1038/nrrheum.2014.144. PMID 25155581.
- ↑ Cohen-Solal JF, Jeganathan V, Grimaldi CM, Peeva E, Diamond B (2006). "Sex hormones and SLE: influencing the fate of autoreactive B cells". Curr. Top. Microbiol. Immunol. 305: 67–88. PMID 16724801.
- ↑ Sullivan KE (2000). "Genetics of systemic lupus erythematosus. Clinical implications". Rheum. Dis. Clin. North Am. 26 (2): 229–56, v–vi. PMID 10768211.
- ↑ Lee HS, Chung YH, Kim TG, Kim TH, Jun JB, Jung S, Bae SC, Yoo DH (2003). "Independent association of HLA-DR and FCgamma receptor polymorphisms in Korean patients with systemic lupus erythematosus". Rheumatology (Oxford). 42 (12): 1501–7. doi:10.1093/rheumatology/keg404. PMID 12867584.
- ↑ 12.0 12.1 Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ (2000). "Systemic lupus erythematosus, complement deficiency, and apoptosis". Adv. Immunol. 76: 227–324. PMID 11079100.
- ↑ Löfgren SE, Yin H, Delgado-Vega AM, Sanchez E, Lewén S, Pons-Estel BA, Witte T, D'Alfonso S, Ortego-Centeno N, Martin J, Alarcón-Riquelme ME, Kozyrev SV (2010). "Promoter insertion/deletion in the IRF5 gene is highly associated with susceptibility to systemic lupus erythematosus in distinct populations, but exerts a modest effect on gene expression in peripheral blood mononuclear cells". J. Rheumatol. 37 (3): 574–8. doi:10.3899/jrheum.090440. PMID 20080916.
- ↑ Sigurdsson S, Nordmark G, Garnier S, Grundberg E, Kwan T, Nilsson O, Eloranta ML, Gunnarsson I, Svenungsson E, Sturfelt G, Bengtsson AA, Jönsen A, Truedsson L, Rantapää-Dahlqvist S, Eriksson C, Alm G, Göring HH, Pastinen T, Syvänen AC, Rönnblom L (2008). "A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5". Hum. Mol. Genet. 17 (18): 2868–76. doi:10.1093/hmg/ddn184. PMC 2525501. PMID 18579578.
- ↑ Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB (2009). "Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo". J. Immunol. 182 (1): 34–8. PMC 2716754. PMID 19109131.
- ↑ Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, Chung SA, Nititham J, Hom G, Kao AH, Demirci FY, Kamboh MI, Petri M, Manzi S, Kastner DL, Seldin MF, Gregersen PK, Behrens TW, Criswell LA (2008). "Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus". PLoS Genet. 4 (5): e1000084. doi:10.1371/journal.pgen.1000084. PMC 2377340. PMID 18516230.
- ↑ Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, Matsumoto I, Ito S, Tsutsumi A, Koga M, Arinami T, Graham RR, Hom G, Takasaki Y, Hashimoto H, Behrens TW, Sumida T, Tsuchiya N (2008). "Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region". Arthritis Res. Ther. 10 (5): R113. doi:10.1186/ar2516. PMC 2592800. PMID 18803832.
- ↑ Yap SN, Phipps ME, Manivasagar M, Tan SY, Bosco JJ (1999). "Human Fc gamma receptor IIA (FcgammaRIIA) genotyping and association with systemic lupus erythematosus (SLE) in Chinese and Malays in Malaysia". Lupus. 8 (4): 305–10. doi:10.1191/096120399678847876. PMID 10413210.
- ↑ Ito I, Kawasaki A, Ito S, Hayashi T, Goto D, Matsumoto I, Tsutsumi A, Hom G, Graham RR, Takasaki Y, Hashimoto H, Ohashi J, Behrens TW, Sumida T, Tsuchiya N (2009). "Replication of the association between the C8orf13-BLK region and systemic lupus erythematosus in a Japanese population". Arthritis Rheum. 60 (2): 553–8. doi:10.1002/art.24246. PMID 19180478.
- ↑ Gregersen PK, Olsson LM (2009). "Recent advances in the genetics of autoimmune disease". Annu. Rev. Immunol. 27: 363–91. doi:10.1146/annurev.immunol.021908.132653. PMC 2992886. PMID 19302045.
- ↑ Yokoyama K, Su Ih IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, Yamamoto T (2002). "BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor". EMBO J. 21 (1–2): 83–92. doi:10.1093/emboj/21.1.83. PMC 125810. PMID 11782428.
- ↑ Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barizzone N, Barrizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, González-Escribano MF, Martin J, Abderrahim H, Alarcón-Riquelme ME (2008). "Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus". Nat. Genet. 40 (2): 211–6. doi:10.1038/ng.79. PMID 18204447.
- ↑ Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA (2008). "Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci". Nat. Genet. 40 (2): 204–10. doi:10.1038/ng.81. PMC 3712260. PMID 18204446.
- ↑ Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC (2007). "Ets-1 is a negative regulator of Th17 differentiation". J. Exp. Med. 204 (12): 2825–35. doi:10.1084/jem.20070994. PMC 2118518. PMID 17967903.
- ↑ 25.0 25.1 Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jönsen A, Bengtsson AA, Rantapää-Dahlqvist S, Baechler EC, Brown EE, Alarcón GS, Edberg JC, Ramsey-Goldman R, McGwin G, Reveille JD, Vilá LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Rönnblom L, Criswell LA, Syvänen AC, Behrens TW, Graham RR (2009). "A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus". Nat. Genet. 41 (11): 1228–33. doi:10.1038/ng.468. PMC 2925843. PMID 19838195.
- ↑ Wojcik H, Griffiths E, Staggs S, Hagman J, Winandy S (2007). "Expression of a non-DNA-binding Ikaros isoform exclusively in B cells leads to autoimmunity but not leukemogenesis". Eur. J. Immunol. 37 (4): 1022–32. doi:10.1002/eji.200637026. PMID 17357110.
- ↑ Petry F, Botto M, Holtappels R, Walport MJ, Loos M (2001). "Reconstitution of the complement function in C1q-deficient (C1qa-/-) mice with wild-type bone marrow cells". J. Immunol. 167 (7): 4033–7. PMID 11564823.
- ↑ Li R, Peng H, Chen GM, Feng CC, Zhang YJ, Wen PF, Qiu LJ, Leng RX, Pan HF, Ye DQ (2014). "Association of FCGR2A-R/H131 polymorphism with susceptibility to systemic lupus erythematosus among Asian population: a meta-analysis of 20 studies". Arch. Dermatol. Res. 306 (9): 781–91. doi:10.1007/s00403-014-1483-5. PMID 24997134.
- ↑ Sepehr A, Wenson S, Tahan SR (2010). "Histopathologic manifestations of systemic diseases: the example of cutaneous lupus erythematosus". J. Cutan. Pathol. 37 Suppl 1: 112–24. doi:10.1111/j.1600-0560.2010.01510.x. PMID 20482683.