L-xylulose reductase
| L-xylulose reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| File:1wnt.jpg L-Xylulose reductase tetramer, Human | |||||||||
| Identifiers | |||||||||
| EC number | 1.1.1.10 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| dicarbonyl/L-xylulose reductase | |
|---|---|
| File:Human xylulose reductase holoenzyme.png | |
| Identifiers | |
| Symbol | DCXR |
| Entrez | 51181 |
| HUGO | 18985 |
| OMIM | 608347 |
| RefSeq | NM_016286 |
| UniProt | Q7Z4W1 |
| Other data | |
| EC number | 1.1.1.10 |
| Locus | Chr. 17 q25.3 |
Dicarbonyl/L-xylulose reductase, also known as carbonyl reductase II, is an enzyme that in human is encoded by the DCXR gene located on chromosome 17.
Structure
The DCXR gene encodes a membrane protein that is approximately 34 kDa in size and composed of 224 amino acids. The protein is highly expressed in the kidney and localizes to the cytoplasmic membrane.[1]
Function
DCSR catalyzes the reduction of several L-xylylose as well as a number of pentoses, tetroses, trioses, alpha-dicarbonyl compounds. The enzyme is involved in carbohydrate metabolism, glucose metabolism, the uronate cycle and may play a role in the water absorption and cellular osmoregulation in the proximal renal tubules by producing xylitol.[2]
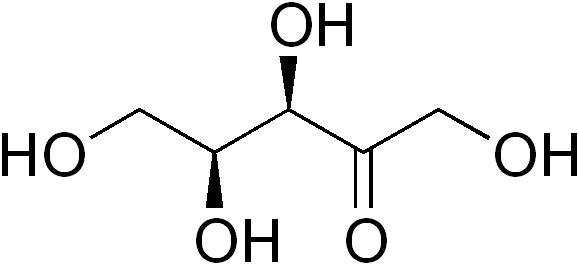
In enzymology, an L-xylulose reductase (EC 1.1.1.10) is an enzyme that catalyzes the chemical reaction
- xylitol + NADP+ <math>\rightleftharpoons</math> L-xylulose + NADPH + H+
Thus, the two substrates of this enzyme are xylitol and NADP+, whereas its 3 products are L-xylulose, NADPH, and H+.
This enzyme belongs to the superfamily of short-chain oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is xylitol:NADP+ 2-oxidoreductase (L-xylulose-forming).
Clinical significance
A deficiency is responsible for pentosuria. The insufficiency of L-xylulose reductase activity causes an inborn error of metabolism disease characterized by excessive urinary excretion of L-xylulose.
Over-expression and ectopic expression of the protein may be associated with prostate adenocarcinoma.[3]
References
- ↑ Nakagawa J, Ishikura S, Asami J, Isaji T, Usami N, Hara A, Sakurai T, Tsuritani K, Oda K, Takahashi M, Yoshimoto M, Otsuka N, Kitamura K (2002). "Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney". J. Biol. Chem. 277 (20): 17883–91. doi:10.1074/jbc.M110703200. PMID 11882650.
- ↑ Zhao HT, Endo S, Ishikura S, Chung R, Hogg PJ, Hara A, El-Kabbani O (2009). "Structure/function analysis of a critical disulfide bond in the active site of L-xylulose reductase". Cell. Mol. Life Sci. 66 (9): 1570–9. doi:10.1007/s00018-009-9065-y. PMID 19337691.
- ↑ Cho-Vega JH, Tsavachidis S, Do KA, Nakagawa J, Medeiros LJ, McDonnell TJ (2007). "Dicarbonyl/L-xylulose reductase: a potential biomarker identified by laser-capture microdissection-micro serial analysis of gene expression of human prostate adenocarcinoma". Cancer Epidemiol. Biomarkers Prev. 16 (12): 2615–22. doi:10.1158/1055-9965.EPI-07-0684. PMID 18086765.
External links
- L-xylulose+reductase at the US National Library of Medicine Medical Subject Headings (MeSH)
| This EC 1.1.1 enzyme-related article is a stub. You can help Wikipedia by expanding it. |