Measles other diagnostic studies
|
Measles Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Measles other diagnostic studies On the Web |
|
American Roentgen Ray Society Images of Measles other diagnostic studies |
|
Risk calculators and risk factors for Measles other diagnostic studies |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Alejandro Lemor, M.D. [2]
Overview
Other diagnostic tests include Vero/hSLAM cells for isolation of measles virus and genetic and sequencing analysis.
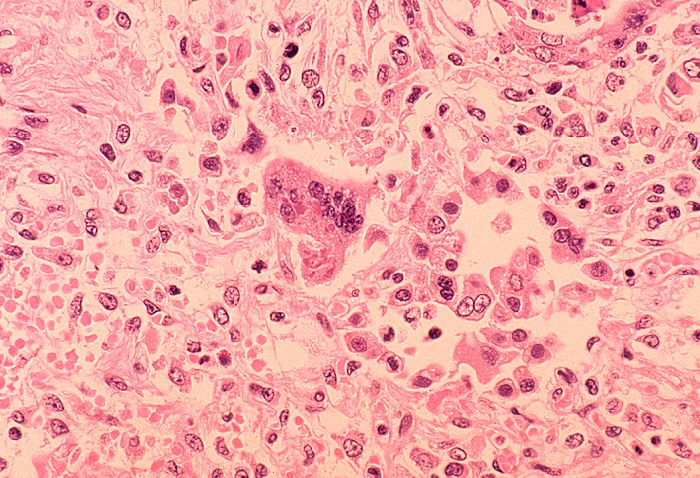
Measles Associated Pneumonia
Some patients will develop pneumonia as a sequela to the measles. Histologically, a unique cell can be found in the paracortical region of hyperplastic lymph nodes in patients affected with this condition. This cell, known as the Warthin-Finkeldey cell, is a multinucleated giant with eosinophilic cytoplasmic and nuclear inclusions. Shown below is an image depicting histological section of lung specimen in a patient with measles associated pneumonia:
Vero/hSLAM Cells for Isolation of Measles Virus[1]
The availability of a sensitive cell line for isolation of measles virus from clinical specimens and establishment of RT-PCR and automated DNA sequencing techniques have allowed for rapid genetic characterization of a large number of wild-type strains of measles virus. A database of sequence information now makes it possible to use molecular epidemiological techniques to identify the source of wild-type viruses and to differentiate between wild type and vaccine strains.
The Vero/hSLAM cell line [hSLAM=(human) signaling lymphocytic activation molecule] is recommended for use in the laboratories that provide measles surveillance as part of the World Health Organization's Measles and Rubella Laboratory Network (WHO LabNet).[2] The Vero/hSLAM cells are Vero cells that have been stably transfected with a plasmid encoding the gene for the human (h) SLAM molecule. SLAM (signaling lymphocyte activation molecule) has been shown to be a receptor for both wild-type and laboratory-adapted strains of measles. The sensitivity of Vero/hSLAM cells for isolation of measles virus is equivalent to that of B95a cells. The advantage to the Vero/hSLAM cells is that, unlike the B95a cells, they are not persistently infected with virus. This provides a significant safety advantage for laboratorians and greatly facilitates international shipments. The disadvantage of the Vero/hSLAM cells is that they must be cultured in medium containing geneticin to retain SLAM expression. This increases the cost of the tissue culture medium.
Genetic Analysis of Measles Viruses
Molecular epidemiology of measles viruses is an important component in outbreak investigations and for global surveillance of circulating wild-type measles strains. The vaccine strain of measles virus can be distinguished from wild-type viruses by determination of the genotype from clinical samples or virus isolates.
Genetic Characterization and Sequencing
Wild-type measles viruses have been divided into distinct genetic groups, referred to as genotypes, based on the nucleotide sequences of their hemagglutinin (H) and nucleoprotein (N) genes, which are the most variable genes on the viral genome.
The 450 nucleotides encoding the carboxy-terminal 150 amino acids of the nucleoprotein has up to 12% nucleotide variation between genotypes. The 450 nucleotides that encode the carboxy-terminal region of the nucleoprotein (N–450) are required for determination of the genotype.
For each genotype, a reference strain is designated for use in genetic analysis (phylogenetic analysis), usually the earliest known virus isolation of that group. The means of referring to the genotypes has been standardized using alphabetical designations for the main groupings (clades). Within the main clades, numerals are added to identify the individual genotypes.
The following 19 genotypes have been detected since 1990: A, B2, B3, C1, C2, D2, D3, D4, D5, D6, D7, D8, D9, D10, D11, G2, G3, H1, H2. During 2011, 8 genotypes were identified by global surveillance: B2, B3, D4, D8, D9, D11, G3, H1
References
- ↑ "CDC Measles Laboratory Findings".
- ↑ Featherstone, D. A.; Rota, P. A.; Icenogle, J.; Mulders, M. N.; Jee, Y.; Ahmed, H.; Bispo de Filippis, A. M.; Ramamurty, N.; Gavrilin, E.; Byabamazima, C.; Dosseh, A.; Xu, W.; Komase, K.; Tashiro, M.; Brown, D.; Bellini, W. J.; Strebel, P. (2011). "Expansion of the Global Measles and Rubella Laboratory Network 2005-09". Journal of Infectious Diseases. 204 (Supplement 1): S491–S498. doi:10.1093/infdis/jir107. ISSN 0022-1899.