Uroporphyrinogen III decarboxylase: Difference between revisions
m (Bot: HTTP→HTTPS) |
imported>MiPe (→Mechanism: typo) |
||
| Line 5: | Line 5: | ||
Uroporphyrinogen III decarboxylase is a homodimeric enzyme ({{PDB|1URO}}) that catalyzes the fifth step in [[heme]] biosynthesis, which corresponds to the elimination of [[carboxyl]] groups from the four [[acetate]] side chains of [[uroporphyrinogen III]] to yield [[coproporphyrinogen III]]: | Uroporphyrinogen III decarboxylase is a homodimeric enzyme ({{PDB|1URO}}) that catalyzes the fifth step in [[heme]] biosynthesis, which corresponds to the elimination of [[carboxyl]] groups from the four [[acetate]] side chains of [[uroporphyrinogen III]] to yield [[coproporphyrinogen III]]: | ||
:[[uroporphyrinogen III]] <math>\rightleftharpoons</math> [[coproporphyrinogen III]] + 4 | :[[uroporphyrinogen III]] <math>\rightleftharpoons</math> [[coproporphyrinogen III]] + 4 CO<sub>2</sub> | ||
== Clinical significance == | == Clinical significance == | ||
| Line 18: | Line 18: | ||
UroD is regarded as an unusual decarboxylase, since it performs decarboxylations without the intervention of any cofactors, unlike the vast majority of decarboxylases. Its mechanism has recently been proposed to proceed through substrate protonation by an [[arginine]] residue.<ref>Silva PJ, Ramos MJ. Density-functional study of mechanisms for the cofactor-free decarboxylation performed by uroporphyrinogen III decarboxylase. ''J Phys Chem B'' 2005;109:18195-200. {{DOI|10.1021/jp051792s}}.</ref> A 2008 report demonstrated that the uncatalyzed rate for UroD's reaction is 10<sup>−19</sup> s<sup>−1</sup>, so at pH 10 the rate acceleration of UroD relative to the uncatalyzed rate, i.e. catalytic proficiency, is the largest for any enzyme known, 6 x 10<sup>24</sup> M<sup>−1</sup>.<ref name="pmid18988736">{{cite journal |vauthors=Lewis CA, Wolfenden R | title = Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 105 | issue = 45 | pages = 17328–33 |date=November 2008 | pmid = 18988736 | doi = 10.1073/pnas.0809838105 | url = | pmc = 2582308 }}</ref> | UroD is regarded as an unusual decarboxylase, since it performs decarboxylations without the intervention of any cofactors, unlike the vast majority of decarboxylases. Its mechanism has recently been proposed to proceed through substrate protonation by an [[arginine]] residue.<ref>Silva PJ, Ramos MJ. Density-functional study of mechanisms for the cofactor-free decarboxylation performed by uroporphyrinogen III decarboxylase. ''J Phys Chem B'' 2005;109:18195-200. {{DOI|10.1021/jp051792s}}.</ref> A 2008 report demonstrated that the uncatalyzed rate for UroD's reaction is 10<sup>−19</sup> s<sup>−1</sup>, so at pH 10 the rate acceleration of UroD relative to the uncatalyzed rate, i.e. catalytic proficiency, is the largest for any enzyme known, 6 x 10<sup>24</sup> M<sup>−1</sup>.<ref name="pmid18988736">{{cite journal |vauthors=Lewis CA, Wolfenden R | title = Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 105 | issue = 45 | pages = 17328–33 |date=November 2008 | pmid = 18988736 | doi = 10.1073/pnas.0809838105 | url = | pmc = 2582308 }}</ref> | ||
[[Image:UroD mechanism.svg|thumb|670x670px|Proposed reaction mechanism of uroporphyrinogen III | [[Image:UroD mechanism.svg|thumb|670x670px|Proposed reaction mechanism of uroporphyrinogen III decarboxylase|center]] | ||
{{Clear}} | {{Clear}} | ||
| Line 26: | Line 26: | ||
==Further reading== | ==Further reading== | ||
{{refbegin | | {{refbegin | 30em}} | ||
{{PBB_Further_reading | {{PBB_Further_reading | ||
| citations = | | citations = | ||
Latest revision as of 09:58, 21 November 2018
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
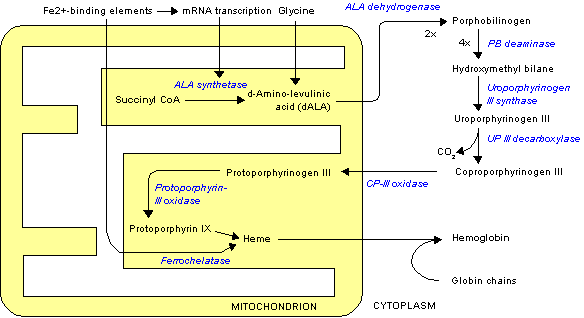
Uroporphyrinogen III decarboxylase (uroporphyrinogen decarboxylase, or UROD) is an enzyme (EC 4.1.1.37) that in humans is encoded by the UROD gene.[1]
Function
Uroporphyrinogen III decarboxylase is a homodimeric enzyme (PDB: 1URO) that catalyzes the fifth step in heme biosynthesis, which corresponds to the elimination of carboxyl groups from the four acetate side chains of uroporphyrinogen III to yield coproporphyrinogen III:
- uroporphyrinogen III <math>\rightleftharpoons</math> coproporphyrinogen III + 4 CO2
Clinical significance
Mutations and deficiency in this enzyme are known to cause familial porphyria cutanea tarda and hepatoerythropoietic porphyria.[1]
Mechanism
At low substrate concentrations, the reaction is believed to follow an ordered route, with the sequential removal of CO2 from the D, A, B, and C rings, whereas at higher substrate/enzyme levels a random route seems to be operative. The enzyme functions as a dimer in solution, and both the enzymes from human and tobacco have been crystallized and solved at good resolutions.
UroD is regarded as an unusual decarboxylase, since it performs decarboxylations without the intervention of any cofactors, unlike the vast majority of decarboxylases. Its mechanism has recently been proposed to proceed through substrate protonation by an arginine residue.[2] A 2008 report demonstrated that the uncatalyzed rate for UroD's reaction is 10−19 s−1, so at pH 10 the rate acceleration of UroD relative to the uncatalyzed rate, i.e. catalytic proficiency, is the largest for any enzyme known, 6 x 1024 M−1.[3]
References
- ↑ 1.0 1.1 "Entrez Gene: UROD uroporphyrinogen decarboxylase".
- ↑ Silva PJ, Ramos MJ. Density-functional study of mechanisms for the cofactor-free decarboxylation performed by uroporphyrinogen III decarboxylase. J Phys Chem B 2005;109:18195-200. doi:10.1021/jp051792s.
- ↑ Lewis CA, Wolfenden R (November 2008). "Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes". Proc. Natl. Acad. Sci. U.S.A. 105 (45): 17328–33. doi:10.1073/pnas.0809838105. PMC 2582308. PMID 18988736.
Further reading
- Elder GH, Lee GB, Tovey JA (1978). "Decreased activity of hepatic uroporphyrinogen decarboxylase in sporadic porphyria cutanea tarda". N. Engl. J. Med. 299 (6): 274–8. doi:10.1056/NEJM197808102990603. PMID 661926.
- de Verneuil H, Bourgeois F, de Rooij F, et al. (1992). "Characterization of a new mutation (R292G) and a deletion at the human uroporphyrinogen decarboxylase locus in two patients with hepatoerythropoietic porphyria". Hum. Genet. 89 (5): 548–52. doi:10.1007/bf00219182. PMID 1634232.
- Romana M, Grandchamp B, Dubart A, et al. (1991). "Identification of a new mutation responsible for hepatoerythropoietic porphyria". Eur. J. Clin. Invest. 21 (2): 225–9. doi:10.1111/j.1365-2362.1991.tb01814.x. PMID 1905636.
- Garey JR, Harrison LM, Franklin KF, et al. (1990). "Uroporphyrinogen decarboxylase: a splice site mutation causes the deletion of exon 6 in multiple families with porphyria cutanea tarda". J. Clin. Invest. 86 (5): 1416–22. doi:10.1172/JCI114856. PMC 296884. PMID 2243121.
- Garey JR, Hansen JL, Harrison LM, et al. (1989). "A point mutation in the coding region of uroporphyrinogen decarboxylase associated with familial porphyria cutanea tarda". Blood. 73 (4): 892–5. PMID 2920211.
- Roméo PH, Raich N, Dubart A, et al. (1986). "Molecular cloning and nucleotide sequence of a complete human uroporphyrinogen decarboxylase cDNA". J. Biol. Chem. 261 (21): 9825–31. PMID 3015909.
- Dubart A, Mattei MG, Raich N, et al. (1986). "Assignment of human uroporphyrinogen decarboxylase (URO-D) to the p34 band of chromosome 1". Hum. Genet. 73 (3): 277–9. doi:10.1007/BF00401245. PMID 3460962.
- Romana M, Dubart A, Beaupain D, et al. (1987). "Structure of the gene for human uroporphyrinogen decarboxylase". Nucleic Acids Res. 15 (18): 7343–56. doi:10.1093/nar/15.18.7343. PMC 306252. PMID 3658695.
- de Verneuil H, Grandchamp B, Beaumont C, et al. (1986). "Uroporphyrinogen decarboxylase structural mutant (Gly281----Glu) in a case of porphyria". Science. 234 (4777): 732–4. doi:10.1126/science.3775362. PMID 3775362.
- Roberts AG, Elder GH, De Salamanca RE, et al. (1995). "A mutation (G281E) of the human uroporphyrinogen decarboxylase gene causes both hepatoerythropoietic porphyria and overt familial porphyria cutanea tarda: biochemical and genetic studies on Spanish patients". J. Invest. Dermatol. 104 (4): 500–2. doi:10.1111/1523-1747.ep12605953. PMID 7706766.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Meguro K, Fujita H, Ishida N, et al. (1994). "Molecular defects of uroporphyrinogen decarboxylase in a patient with mild hepatoerythropoietic porphyria". J. Invest. Dermatol. 102 (5): 681–5. doi:10.1111/1523-1747.ep12374134. PMID 8176248.
- Moran-Jimenez MJ, Ged C, Romana M, et al. (1996). "Uroporphyrinogen decarboxylase: complete human gene sequence and molecular study of three families with hepatoerythropoietic porphyria". Am. J. Hum. Genet. 58 (4): 712–21. PMC 1914669. PMID 8644733.
- McManus JF, Begley CG, Sassa S, Ratnaike S (1996). "Five new mutations in the uroporphyrinogen decarboxylase gene identified in families with cutaneous porphyria". Blood. 88 (9): 3589–600. PMID 8896428.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Whitby FG, Phillips JD, Kushner JP, Hill CP (1998). "Crystal structure of human uroporphyrinogen decarboxylase". EMBO J. 17 (9): 2463–71. doi:10.1093/emboj/17.9.2463. PMC 1170588. PMID 9564029.
- Mendez M, Sorkin L, Rossetti MV, et al. (1998). "Familial porphyria cutanea tarda: characterization of seven novel uroporphyrinogen decarboxylase mutations and frequency of common hemochromatosis alleles". Am. J. Hum. Genet. 63 (5): 1363–75. doi:10.1086/302119. PMC 1377546. PMID 9792863.
- Wang H, Long Q, Marty SD, et al. (1998). "A zebrafish model for hepatoerythropoietic porphyria". Nat. Genet. 20 (3): 239–43. doi:10.1038/3041. PMID 9806541.
- McManus JF, Begley CG, Sassa S, Ratnaike S (1999). "Three new mutations in the uroporphyrinogen decarboxylase gene in familial porphyria cutanea tarda. Mutation in brief no. 237. Online". Hum. Mutat. 13 (5): 412–413. doi:10.1002/(SICI)1098-1004(1999)13:5<412::AID-HUMU13>3.0.CO;2-N. PMID 10338097.
- Christiansen L, Ged C, Hombrados I, et al. (1999). "Screening for mutations in the uroporphyrinogen decarboxylase gene using denaturing gradient gel electrophoresis. Identification and characterization of six novel mutations associated with familial PCT". Hum. Mutat. 14 (3): 222–32. doi:10.1002/(SICI)1098-1004(1999)14:3<222::AID-HUMU5>3.0.CO;2-V. PMID 10477430.