Pyridoxal phosphate: Difference between revisions
m (Robot: Automated text replacement (-{{reflist}} +{{reflist|2}}, -<references /> +{{reflist|2}}, -{{WikiDoc Cardiology Network Infobox}} +)) |
Kiran Singh (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
{{ | __Notoc__ | ||
|ImageFile=Pyridoxal-phosphate.svg | {{SI}} | ||
|ImageSize= | {{CMG}} | ||
|IUPACName=(4-formyl-5-hydroxy | ==Overview== | ||
|OtherNames= | {{chembox | ||
|Section1= {{Chembox Identifiers | | Verifiedfields = changed | ||
| | | verifiedrevid = 393759663 | ||
| | | ImageFile_Ref = {{chemboximage|correct|??}} | ||
| | | ImageFile = Pyridoxal-phosphate.svg | ||
| | | ImageSize = | ||
| ImageName = Skeletal formula | |||
| ImageFile1 = Pyridoxal-phosphate-3D-balls.png | |||
| ImageSize1 = 210px | |||
| ImageName1 = Ball-and-stick model | |||
| IUPACName =(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methyl phosphate | |||
| OtherNames = Pyridoxal 5-phosphate, PAL-P, PLP, Vitamin B6 phosphate<ref>Anonymous . Substance Detail. https://scifinder-cas-org.proxy.library.nd.edu:9443/scifinder/view/scifinder/scifinderExplore.jsf (accessed 12 Nov, 2011).</ref> | |||
|Section1={{Chembox Identifiers | |||
| CASNo_Ref = {{cascite|correct|CAS}} | |||
| CASNo =54-47-7 | |||
| PubChem =1051 | |||
| ChEBI = 18405 | |||
| ChEMBL_Ref = {{ebicite|changed|EBI}} | |||
| ChEMBL = 82202 | |||
| SMILES = CC1=NC=C(C(=C1O)C=O)COP(=O)(O)O | |||
| MeSHName =Pyridoxal+Phosphate | |||
| ATCCode_prefix = A11 | |||
| ATCCode_suffix = HA06 | |||
}} | }} | ||
|Section2= {{Chembox Properties | |Section2={{Chembox Properties | ||
| | | Formula =C<sub>8</sub>H<sub>10</sub>NO<sub>6</sub>P | ||
| | | MolarMass =247.142 g/mol | ||
| | | Appearance = | ||
| | | Density =1.638±0.06 g/cm3<ref name="ReferenceA">Calculated using Advanced Chemistry Development (ACD/Labs) Software V11.02 (© 1994-2011 ACD/Labs)</ref> | ||
| MeltingPtC = 139 to 142 | |||
| | | Melting_ref = <ref>Kozlov E.I., L. M. S. Stability of water-soluble vitamins and coenzymes. Hydrolysis of pyridoxal-5-phosphate in acidic, neutral, and weakly alkaline solutions. Pharmaceutical Chemistry Journal 1978, 11, 1543.</ref> | ||
| | | BoilingPt = | ||
| Solubility = | |||
| | | pKa =1.56<ref name="ReferenceA"/> | ||
| | |||
| | |||
}} | }} | ||
|Section3={{Chembox Hazards | |||
| MainHazards = | |||
| FlashPt =296.0±32.9 °C<ref name="ReferenceA"/> | |||
| AutoignitionPt = | |||
}} | |||
}} | }} | ||
{{ | '''Pyridoxal phosphate''' (PLP, [[pyridoxal]] 5'-[[phosphate]], P5P), the active form of [[vitamin B6|vitamin B<sub>6</sub>]], is a [[coenzyme]] in a variety of [[enzymatic]] reactions. The [[Enzyme commission]] has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities.<ref>{{cite journal | author = Percudani R1, Peracchi A. | year = 2003 | title = A genomic overview of pyridoxal-phosphate-dependent enzymes | journal = EMBO Rep. | volume = 4 | issue = 9 | pages = 850–4 | pmid = 12949584 | pmc = 1326353 | doi=10.1038/sj.embor.embor914}}</ref> The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates. | ||
==Etymology== | |||
[[File:Pyridine numbers.svg|100px|thumbnail|Pyridine]] | |||
PLP stands for '''p'''yridoxa'''l''' 5' '''p'''hosphate and PMP stands for '''p'''yridoxa'''m'''ine 5' '''p'''hosphate. Consequently, the L in PLP does not indicate a levorotatory chiral center. | |||
The common names pyridoxamine (aminomethyl group at position 4), pyroxidal (carbaldehyde group) and pyridoxine (alkaloid, has hydroxymethyl) derive from pyridine + oxy + a [[IUPAC nomenclature of organic chemistry|IUPAC suffix]].Pyridine, in turn, comes from the Greek πῦρ, πυρός (pyr, fire) + -ide + -ine. | |||
It should be noted that the oxy infix in pyridoxine is not based on [[Hantzsch–Widman nomenclature]] (the heterocyclic ring is simply a modified pyridine) and the pyr- root does not indicate it to be a dimeric acid anhydride (unlike pyrophosphate). | |||
==Role as a coenzyme== | ==Role as a coenzyme== | ||
PLP acts as a coenzyme in all [[transamination]] reactions, and in | PLP acts as a coenzyme in all [[transamination]] reactions, and in certain [[decarboxylation]], [[deamination]], and [[racemization]] reactions of [[amino acid]]s.<ref name=Dolphin>David Dolphin, R Poulson, and O Avramovic. Vitamin B6: Pyridoxal Phosphate Volume 1, Part B, Coenzymes and Cofactors. Wiley Interscience, New YorkYear: 1986 ISBN 978-0471097853. [http://www.daviddolphin.com/books/book15-volume-preface.pdf Preface].</ref> The aldehyde group of PLP forms a [[Schiff base|Schiff-base]] linkage (internal [[aldimine]]) with the ε-amino group of a specific lysine group of the [[aminotransferase]] enzyme. The α-amino group of the amino acid substrate displaces the ε-amino group of the active-site lysine residue in a process known as transaldimination. The resulting external aldimine can lose a proton, carbon dioxide, or an amino acid sidechain to become a quinoid intermediate, which in turn can act as a nucleophile in several reaction pathways. | ||
In transamination, after deprotonation the quinoid intermediate accepts a proton at a different position to become a [[ketimine]]. The resulting ketimine is hydrolysed so that the amino group remains on the complex.<ref>Toney, M. D. "Reaction specificity in pyridoxal enzymes." Archives of biochemistry and biophysics (2005) 433: 279-287.</ref> In addition, PLP is used by aminotransferases (or transaminases) that act upon unusual sugars such as [[perosamine]] and [[desosamine]].<ref name="samuel">Samuel, G. and Reeves, P. "Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precusor synthesis and O-antigen assembly." Carbohydrate research (2003) 338:2503-2519.</ref> In these reactions, the PLP reacts with [[glutamate]], which transfers its alpha-amino group to PLP to make pyridoxamine phosphate (PMP). PMP then transfers its nitrogen to the sugar, making an [[amino sugar]]. | |||
PLP is also involved in various beta-elimination reactions such as the reactions carried out by [[serine dehydratase]] and [[ColD|GDP-4-keto-6-deoxymannose-3-dehydratase (ColD)]]<ref name="samuel"/> | PLP is also involved in various beta-elimination reactions such as the reactions carried out by [[serine dehydratase]] and [[ColD|GDP-4-keto-6-deoxymannose-3-dehydratase (ColD)]].<ref name="samuel"/> | ||
It is also active in the condensation reaction in [[heme]] synthesis. | It is also active in the condensation reaction in [[heme]] synthesis. | ||
PLP plays a role in the conversion of [[levodopa]] into [[dopamine]], facilitates the conversion of the excitatory neurotransmitter glutamate to the inhibitory neurotransmitter [[GABA]], and allows [[S-adenosyl methionine|SAM]] to be decarboxylated to form [[propylamine]], which is a precursor to polyamines. | |||
==Non-classical examples of PLP== | ==Non-classical examples of PLP== | ||
PLP is also found on glycogen phosphorylase in the liver, where it is used to break down [[glycogen]] in [[ | PLP is also found on [[glycogen phosphorylase]] in the liver, where it is used to break down [[glycogen]] in [[glycogenolysis]] when [[glucagon]] or [[epinephrine]] signals it to do so. However, this enzyme does not exploit the reactive aldehyde group, but instead utilizes the phosphate group on PLP to perform its reaction. | ||
Although the vast majority of PLP-dependent enzymes form an internal aldimine with PLP via an active site lysine residue, some PLP-dependent enzymes do not have this lysine residue, but instead have a histidine in the active site. In such a case, the histidine cannot form the internal aldimine, and, therefore, the co-factor does not become covalently tethered to the enzyme. [[ColD|GDP-4-keto-6-deoxymannose-3-dehydratase (ColD)]] is an example of such an enzyme.<ref>Cook P. D., Thoden J.B. and Holden H. M. "The structure of GDP-4-keto-6-deoxymannose-3-dehydratase: a unique coenzyme B6-dependent enzyme." Protein Science (2006) 15:2093-2106. | |||
</ref> | |||
==Catalytic mechanism== | |||
The pyridoxal-5′-phosphate-dependent enzymes (PLP enzymes) catalyze a myriad of biochemical reactions. Although the scope of PLP-catalyzed reactions appears to be immensely diverse, there is a simple unifying principle: in the resting state, the co-factor (PLP) is covalently bonded to the amino group of an active site lysine, forming an internal aldimine. Once the amino substrate interacts with the active site, a new Schiff base is generated, commonly referred to as the external aldimine. After this step, the mechanistic pathway for each PLP-catalyzed reaction diverges. Density functional methods have been applied to investigate the transimination reaction, and the results have shown that the reaction involves three sequential steps: (i) formation of a tetrahedral intermediate with the active site lysine and the amino substrate bonded to the PLP co-factor; (ii) non-direct proton transfer between the amino substrate and the lysine residue; and (iii) formation of the external aldimine after the dissociation of the lysine residue. The overall reaction is exothermic (−12.0 kcal/mol), the second step being rate-limiting, with 12.6 kcal/mol for the activation energy<ref>{{cite journal | author = N. M. F. S. A. Cerqueira, P. A. Fernandes, M. J. Ramos | title = Computational Mechanistic Studies Addressed to the Transimination Reaction Present in All Pyridoxal 5′-Phosphate-Requiring Enzymes | journal = [[Journal of Chemical Theory and Computation]] | year = 2011 | volume = 7 | issue = 5 | pages = 1356–1368 | doi = 10.1021/ct1002219}}</ref> | |||
[[File:PLP mechanism.svg|800px|thumbnail|center|Mechanistic examples: racemization of alanine and elimination of cysteine.]] | |||
==Specificity== | |||
Specificity is conferred by the fact that, of the four bonds of the alpha-carbon of the amino acid aldimine state, the bond perpendicular to the pyridine ring will be broken ([[Dunathan Stereoelectronic Hypothesis]]).<ref name=rev>{{cite doi|10.1146/annurev.biochem.73.011303.074021}}</ref> Consequently, specificity is dictated by how the enzymes bind their substrates. | |||
An additional role in specificity is played by the ease of protonation of the [[pyridine]] ring nitrogen.<ref>{{cite pmid|21827189}}</ref> | |||
==PLP-enzymes== | |||
PLP is retained in the active site not only thanks to the lysine, but also thanks to the interaction of the phosphate group and a phosphate binding pocket and to a lesser extent thanks to base stacking of the pyridine ring with an overhanging aromatic residue, generally tyrosine (which may also partake in the acid–base catalysis). Despite the limited requirements for a PLP binding pocket, PLP enzymes belong to only five different families. These families do not correlate well with a particular type of reaction. The five families are classified as fold types followed by a Roman numeral.<ref name=rev/> | |||
* Fold Type I — aspartate aminotransferase family | |||
* Fold Type II — tryptophan synthase family | |||
* Fold Type IV — D-amino acid aminotransferase family | |||
* Fold Type III — alanine racemase family (TIM-barrel) | |||
* Fold Type V — glycogen phosphorylase family | |||
==Biosynthesis== | |||
===From vitamers=== | |||
Animals are auxotrophs for this enzyme co-factor and require it or an intermediate to be supplemented, hence its classification as a vitamin B<sub>6</sub>, unlike [[Molybdenum cofactor|MoCo]] or [[CoQ10]] for example. PLP is synthesized from pyridoxal by the enzyme [[pyridoxal kinase]], requiring one ATP molecule. PLP is metabolized in the liver. | |||
===Prototrophy=== | |||
Two natural pathways for PLP are currently known: one requires deoxyxylulose 5-phosphate (DXP), while the other does not, hence they are known as DXP-dependent and DXP-independent. These pathways have been studied extensively in ''Escherichia coli'' and ''Bacillus subtilis'', respectively. Despite the disparity in the starting compounds and the different number of steps required, the two pathways possess many commonalities.<ref>{{cite pmid|17822383}}</ref> | |||
====DXP-dependent biosynthesis==== | |||
The DXP-dependent biosynthetic route requires several steps and a convergence of two branches, one producing [[3-hydroxy-1-aminoacetone phosphate]] from [[erythrose 4-phosphate]], while the other (single enzyme) producing [[deoxyxylulose 5-phosphate]] (DXP) from [[glyceraldehyde 3-phosphate]] (GAP) and [[pyruvate]]. The condensation product of [[3-hydroxy-1-aminoacetone phosphate]] and [[deoxyxylulose 5-phosphate]] is [[pyridoxine 5'-phosphate]]. The condensation is catalyzed by [[PNP synthase]], encoded by ''pdxJ'', which creates PNP (pyridoxine 5' phosphate).<ref>{{cite pmid|15242009}}</ref> The final enzyme is PNP oxidase (''pdxH''), which catalyzes the oxidation of the 4' hydroxyl group to an aldehyde using dioxigen, resulting in hydrogen peroxide. | |||
The first branch is catalyzed in ''E. coli'' by enzymes encoded by ''epd'', ''pdxB'', ''serC'' and ''pdxA''. These share mechanistical similarities and homology with the three enzymes in serine biosynthesis (''serA'' (homologue of ''pdxB''), ''serC'', ''serB'' &emdash;however, ''epd'' is a homologue of ''gap''), which points towards a shared evolutionary origin of the two pathways.<ref>{{cite pmid|2121717}}</ref> In several species there are two homologues of the ''E. coli'' ''serC'' gene, generally one in a ser operon (''serC''), and the other in a pdx operon, in which case it is called ''pdxF''. | |||
[[File:Metabolic pathway- pyridoxal 5'-phosphate biosynthesis I v 2.0.svg|700px|center]] | |||
A "serendipitous pathway" was found in an overexpression library that could suppress the auxotrophy caused by the deletion of pdxB (encoding erythronate 4 phosphate dehydrogenase) in ''E. coli''. The serendipitous pathway was very inefficient, but was possible due to the [[enzyme promiscuity|promiscuous activity]] of various enzymes. It started with 3-phosphohydroxypyruvate (the product of the ''serA''-encoded enzyme in serine biosynthesis) and did not require erythronate-4-phosphate. 3PHP was dephosphorylated, resulting in an unstable intermediate that decarboxylates spontaneously (hence the presence of the phosphate in the serine biosynthetic pathway) to glycaldehyde. Glycaldehyde was condensed with glycine and the phosphorylated product was 4-phosphohydroxythreonine (4PHT), the canonical substate for 4-PHT dehydrogenase (''pdxA'').<ref>{{cite doi|10.1038/msb.2010.88}}</ref> | |||
====DXP-independent biosynthesis==== | |||
The DXP-independent PLP-biosynthetic route consists of a step catalyzed by PLP-synthase, an enzyme composed of two subunits. PdxS catalyzes the condensation of ribulose 5-phosphate, glyceraldehyde-3-phosphate, and [[ammonia]], this latter molecules is produced by PdxT which catalyzes the production of ammonia from [[glutamine]]. PdxS is a (β/α)8 barrel (also known as a TIM-barrel) that forms a dodecamer.<ref>{{cite pmid|15911615}}</ref> | |||
===Prebiotic synthesis=== | |||
The widespread utilization of PLP in central metabolism, especially in amino acid biosynthesis, and its activity in the absence of enzymes, suggests PLP may be a prebiotic compound.<ref name=prebio99>{{cite pmid|10389266}}</ref> | |||
In fact, heating NH3 and glycoaldehyde spontaneously forms a variety of pyridines, including pyridoxal.<ref name=prebio99/> Under certain conditions, PLP is formed from cyanoacetylene, diacetylene, carbon monoxide, hydrogen, water, and a phosphoric acid.<ref>{{cite pmid|16730878}}</ref> | |||
==Inhibitors== | |||
Several inhibitors of PLP enzymes are known. | |||
One type of inhibitor forms an electrophile with PLP, causing it to irreversibly react with the active site lysine. Acetylenic compounds (e.g. propargylglycine) and vinylic compounds (e.g. vinylglycine) are such inhibitors. | |||
A different type of inhibitor inactivates PLP, and such are α-methyl and amino-oxy substrate analogs (e.g. α-methylglutamate). | |||
Other inhibitors have good leaving groups that nucleophilically attack the PLP. Such is chloroalanine, which inhibits a large number of enzymes.<ref name=rev/> | |||
Examples of inhibitors: | |||
* AlaP (alanine phosphonate) inihibits [[alanine racemase]]s, but its lack of specificity has prompted further designs of ALR inhibitors.<ref>{{cite pmid|21637807}}</ref> | |||
* [[Gabaculine]] and [[Vigabatrin]] inhibit [[GABA aminotransferase]]{{disambiguation needed|date=May 2013}} | |||
* [[Canaline]] and 5-fluoromethylornithine inhibit [[ornithine aminotransferase]] | |||
* Amino-oxy SAM inhibits [[1-aminocyclopropane-1-carboxylate synthase|ACC synthase]] | |||
== See also == | == See also == | ||
| Line 58: | Line 131: | ||
==References== | ==References== | ||
{{reflist | {{reflist}} | ||
==External links== | ==External links== | ||
* {{ATC|A11|HA06}} | * {{ATC|A11|HA06}} | ||
{{Vitamin}} | {{Vitamin}} | ||
{{Enzyme cofactors}} | {{Enzyme cofactors}} | ||
{{ | {{DEFAULTSORT:Pyridoxal Phosphate}} | ||
[[Category:B vitamins]] | |||
[[Category:Phosphates]] | |||
[[ru:Витамин B6#Пиридоксальфосфат]] | |||
Revision as of 13:30, 7 April 2015
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Template:Chembox E numberTemplate:Chembox DensityTemplate:Chembox MeltingPtTemplate:Chembox pKaTemplate:Chembox FlashPt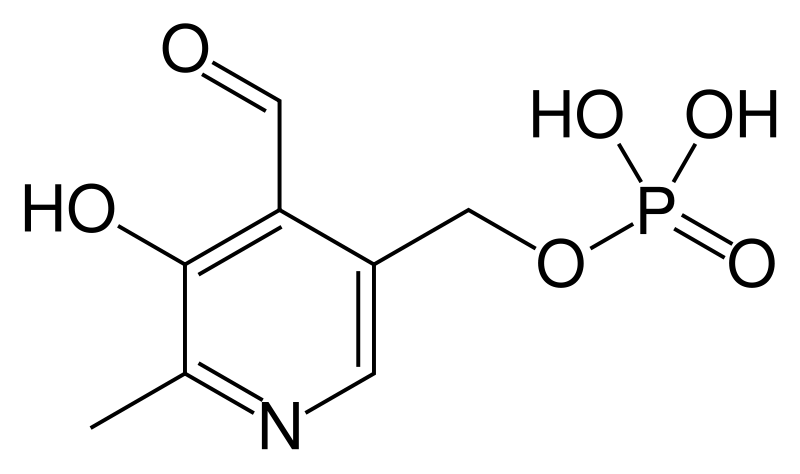
| |
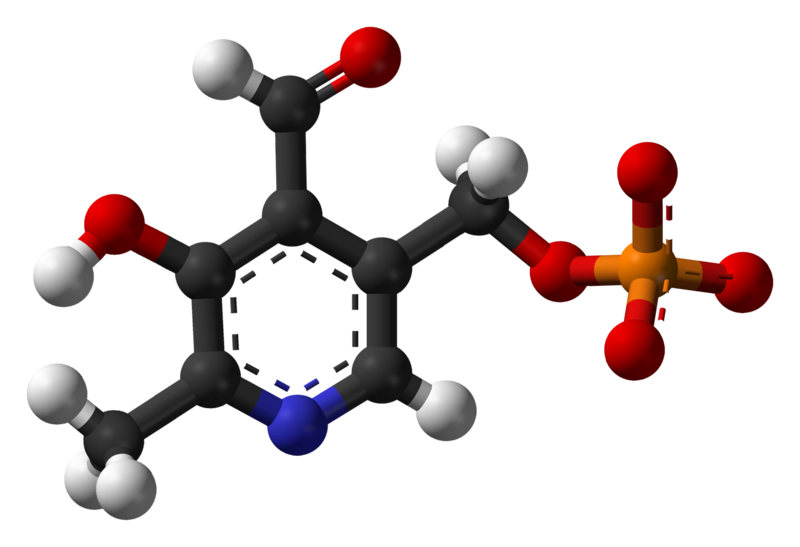
| |
| Names | |
|---|---|
| IUPAC name
(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methyl phosphate
| |
| Other names
Pyridoxal 5-phosphate, PAL-P, PLP, Vitamin B6 phosphate[1]
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ECHA InfoCard | Lua error in Module:Wikidata at line 879: attempt to index field 'wikibase' (a nil value). Lua error in Module:Wikidata at line 879: attempt to index field 'wikibase' (a nil value). |
| MeSH | Pyridoxal+Phosphate |
PubChem CID
|
|
| |
| Properties | |
| C8H10NO6P | |
| Molar mass | 247.142 g/mol |
| Hazards | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The Enzyme commission has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities.[4] The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Etymology
PLP stands for pyridoxal 5' phosphate and PMP stands for pyridoxamine 5' phosphate. Consequently, the L in PLP does not indicate a levorotatory chiral center.
The common names pyridoxamine (aminomethyl group at position 4), pyroxidal (carbaldehyde group) and pyridoxine (alkaloid, has hydroxymethyl) derive from pyridine + oxy + a IUPAC suffix.Pyridine, in turn, comes from the Greek πῦρ, πυρός (pyr, fire) + -ide + -ine. It should be noted that the oxy infix in pyridoxine is not based on Hantzsch–Widman nomenclature (the heterocyclic ring is simply a modified pyridine) and the pyr- root does not indicate it to be a dimeric acid anhydride (unlike pyrophosphate).
Role as a coenzyme
PLP acts as a coenzyme in all transamination reactions, and in certain decarboxylation, deamination, and racemization reactions of amino acids.[5] The aldehyde group of PLP forms a Schiff-base linkage (internal aldimine) with the ε-amino group of a specific lysine group of the aminotransferase enzyme. The α-amino group of the amino acid substrate displaces the ε-amino group of the active-site lysine residue in a process known as transaldimination. The resulting external aldimine can lose a proton, carbon dioxide, or an amino acid sidechain to become a quinoid intermediate, which in turn can act as a nucleophile in several reaction pathways.
In transamination, after deprotonation the quinoid intermediate accepts a proton at a different position to become a ketimine. The resulting ketimine is hydrolysed so that the amino group remains on the complex.[6] In addition, PLP is used by aminotransferases (or transaminases) that act upon unusual sugars such as perosamine and desosamine.[7] In these reactions, the PLP reacts with glutamate, which transfers its alpha-amino group to PLP to make pyridoxamine phosphate (PMP). PMP then transfers its nitrogen to the sugar, making an amino sugar.
PLP is also involved in various beta-elimination reactions such as the reactions carried out by serine dehydratase and GDP-4-keto-6-deoxymannose-3-dehydratase (ColD).[7]
It is also active in the condensation reaction in heme synthesis.
PLP plays a role in the conversion of levodopa into dopamine, facilitates the conversion of the excitatory neurotransmitter glutamate to the inhibitory neurotransmitter GABA, and allows SAM to be decarboxylated to form propylamine, which is a precursor to polyamines.
Non-classical examples of PLP
PLP is also found on glycogen phosphorylase in the liver, where it is used to break down glycogen in glycogenolysis when glucagon or epinephrine signals it to do so. However, this enzyme does not exploit the reactive aldehyde group, but instead utilizes the phosphate group on PLP to perform its reaction.
Although the vast majority of PLP-dependent enzymes form an internal aldimine with PLP via an active site lysine residue, some PLP-dependent enzymes do not have this lysine residue, but instead have a histidine in the active site. In such a case, the histidine cannot form the internal aldimine, and, therefore, the co-factor does not become covalently tethered to the enzyme. GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) is an example of such an enzyme.[8]
Catalytic mechanism
The pyridoxal-5′-phosphate-dependent enzymes (PLP enzymes) catalyze a myriad of biochemical reactions. Although the scope of PLP-catalyzed reactions appears to be immensely diverse, there is a simple unifying principle: in the resting state, the co-factor (PLP) is covalently bonded to the amino group of an active site lysine, forming an internal aldimine. Once the amino substrate interacts with the active site, a new Schiff base is generated, commonly referred to as the external aldimine. After this step, the mechanistic pathway for each PLP-catalyzed reaction diverges. Density functional methods have been applied to investigate the transimination reaction, and the results have shown that the reaction involves three sequential steps: (i) formation of a tetrahedral intermediate with the active site lysine and the amino substrate bonded to the PLP co-factor; (ii) non-direct proton transfer between the amino substrate and the lysine residue; and (iii) formation of the external aldimine after the dissociation of the lysine residue. The overall reaction is exothermic (−12.0 kcal/mol), the second step being rate-limiting, with 12.6 kcal/mol for the activation energy[9]
Specificity
Specificity is conferred by the fact that, of the four bonds of the alpha-carbon of the amino acid aldimine state, the bond perpendicular to the pyridine ring will be broken (Dunathan Stereoelectronic Hypothesis).[10] Consequently, specificity is dictated by how the enzymes bind their substrates. An additional role in specificity is played by the ease of protonation of the pyridine ring nitrogen.[11]
PLP-enzymes
PLP is retained in the active site not only thanks to the lysine, but also thanks to the interaction of the phosphate group and a phosphate binding pocket and to a lesser extent thanks to base stacking of the pyridine ring with an overhanging aromatic residue, generally tyrosine (which may also partake in the acid–base catalysis). Despite the limited requirements for a PLP binding pocket, PLP enzymes belong to only five different families. These families do not correlate well with a particular type of reaction. The five families are classified as fold types followed by a Roman numeral.[10]
- Fold Type I — aspartate aminotransferase family
- Fold Type II — tryptophan synthase family
- Fold Type IV — D-amino acid aminotransferase family
- Fold Type III — alanine racemase family (TIM-barrel)
- Fold Type V — glycogen phosphorylase family
Biosynthesis
From vitamers
Animals are auxotrophs for this enzyme co-factor and require it or an intermediate to be supplemented, hence its classification as a vitamin B6, unlike MoCo or CoQ10 for example. PLP is synthesized from pyridoxal by the enzyme pyridoxal kinase, requiring one ATP molecule. PLP is metabolized in the liver.
Prototrophy
Two natural pathways for PLP are currently known: one requires deoxyxylulose 5-phosphate (DXP), while the other does not, hence they are known as DXP-dependent and DXP-independent. These pathways have been studied extensively in Escherichia coli and Bacillus subtilis, respectively. Despite the disparity in the starting compounds and the different number of steps required, the two pathways possess many commonalities.[12]
DXP-dependent biosynthesis
The DXP-dependent biosynthetic route requires several steps and a convergence of two branches, one producing 3-hydroxy-1-aminoacetone phosphate from erythrose 4-phosphate, while the other (single enzyme) producing deoxyxylulose 5-phosphate (DXP) from glyceraldehyde 3-phosphate (GAP) and pyruvate. The condensation product of 3-hydroxy-1-aminoacetone phosphate and deoxyxylulose 5-phosphate is pyridoxine 5'-phosphate. The condensation is catalyzed by PNP synthase, encoded by pdxJ, which creates PNP (pyridoxine 5' phosphate).[13] The final enzyme is PNP oxidase (pdxH), which catalyzes the oxidation of the 4' hydroxyl group to an aldehyde using dioxigen, resulting in hydrogen peroxide.
The first branch is catalyzed in E. coli by enzymes encoded by epd, pdxB, serC and pdxA. These share mechanistical similarities and homology with the three enzymes in serine biosynthesis (serA (homologue of pdxB), serC, serB &emdash;however, epd is a homologue of gap), which points towards a shared evolutionary origin of the two pathways.[14] In several species there are two homologues of the E. coli serC gene, generally one in a ser operon (serC), and the other in a pdx operon, in which case it is called pdxF.
A "serendipitous pathway" was found in an overexpression library that could suppress the auxotrophy caused by the deletion of pdxB (encoding erythronate 4 phosphate dehydrogenase) in E. coli. The serendipitous pathway was very inefficient, but was possible due to the promiscuous activity of various enzymes. It started with 3-phosphohydroxypyruvate (the product of the serA-encoded enzyme in serine biosynthesis) and did not require erythronate-4-phosphate. 3PHP was dephosphorylated, resulting in an unstable intermediate that decarboxylates spontaneously (hence the presence of the phosphate in the serine biosynthetic pathway) to glycaldehyde. Glycaldehyde was condensed with glycine and the phosphorylated product was 4-phosphohydroxythreonine (4PHT), the canonical substate for 4-PHT dehydrogenase (pdxA).[15]
DXP-independent biosynthesis
The DXP-independent PLP-biosynthetic route consists of a step catalyzed by PLP-synthase, an enzyme composed of two subunits. PdxS catalyzes the condensation of ribulose 5-phosphate, glyceraldehyde-3-phosphate, and ammonia, this latter molecules is produced by PdxT which catalyzes the production of ammonia from glutamine. PdxS is a (β/α)8 barrel (also known as a TIM-barrel) that forms a dodecamer.[16]
Prebiotic synthesis
The widespread utilization of PLP in central metabolism, especially in amino acid biosynthesis, and its activity in the absence of enzymes, suggests PLP may be a prebiotic compound.[17] In fact, heating NH3 and glycoaldehyde spontaneously forms a variety of pyridines, including pyridoxal.[17] Under certain conditions, PLP is formed from cyanoacetylene, diacetylene, carbon monoxide, hydrogen, water, and a phosphoric acid.[18]
Inhibitors
Several inhibitors of PLP enzymes are known.
One type of inhibitor forms an electrophile with PLP, causing it to irreversibly react with the active site lysine. Acetylenic compounds (e.g. propargylglycine) and vinylic compounds (e.g. vinylglycine) are such inhibitors. A different type of inhibitor inactivates PLP, and such are α-methyl and amino-oxy substrate analogs (e.g. α-methylglutamate). Other inhibitors have good leaving groups that nucleophilically attack the PLP. Such is chloroalanine, which inhibits a large number of enzymes.[10]
Examples of inhibitors:
- AlaP (alanine phosphonate) inihibits alanine racemases, but its lack of specificity has prompted further designs of ALR inhibitors.[19]
- Gabaculine and Vigabatrin inhibit GABA aminotransferase[disambiguation needed]
- Canaline and 5-fluoromethylornithine inhibit ornithine aminotransferase
- Amino-oxy SAM inhibits ACC synthase
See also
References
- ↑ Anonymous . Substance Detail. https://scifinder-cas-org.proxy.library.nd.edu:9443/scifinder/view/scifinder/scifinderExplore.jsf (accessed 12 Nov, 2011).
- ↑ 2.0 2.1 2.2 Calculated using Advanced Chemistry Development (ACD/Labs) Software V11.02 (© 1994-2011 ACD/Labs)
- ↑ Kozlov E.I., L. M. S. Stability of water-soluble vitamins and coenzymes. Hydrolysis of pyridoxal-5-phosphate in acidic, neutral, and weakly alkaline solutions. Pharmaceutical Chemistry Journal 1978, 11, 1543.
- ↑ Percudani R1, Peracchi A. (2003). "A genomic overview of pyridoxal-phosphate-dependent enzymes". EMBO Rep. 4 (9): 850–4. doi:10.1038/sj.embor.embor914. PMC 1326353. PMID 12949584.
- ↑ David Dolphin, R Poulson, and O Avramovic. Vitamin B6: Pyridoxal Phosphate Volume 1, Part B, Coenzymes and Cofactors. Wiley Interscience, New YorkYear: 1986 ISBN 978-0471097853. Preface.
- ↑ Toney, M. D. "Reaction specificity in pyridoxal enzymes." Archives of biochemistry and biophysics (2005) 433: 279-287.
- ↑ 7.0 7.1 Samuel, G. and Reeves, P. "Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precusor synthesis and O-antigen assembly." Carbohydrate research (2003) 338:2503-2519.
- ↑ Cook P. D., Thoden J.B. and Holden H. M. "The structure of GDP-4-keto-6-deoxymannose-3-dehydratase: a unique coenzyme B6-dependent enzyme." Protein Science (2006) 15:2093-2106.
- ↑ N. M. F. S. A. Cerqueira, P. A. Fernandes, M. J. Ramos (2011). "Computational Mechanistic Studies Addressed to the Transimination Reaction Present in All Pyridoxal 5′-Phosphate-Requiring Enzymes". Journal of Chemical Theory and Computation. 7 (5): 1356–1368. doi:10.1021/ct1002219.
- ↑ 10.0 10.1 10.2 Template:Cite doi
- ↑ PMID 21827189 (PMID 21827189)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ PMID 17822383 (PMID 17822383)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ PMID 15242009 (PMID 15242009)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ PMID 2121717 (PMID 2121717)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ Template:Cite doi
- ↑ PMID 15911615 (PMID 15911615)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ 17.0 17.1 PMID 10389266 (PMID 10389266)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ PMID 16730878 (PMID 16730878)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand - ↑ PMID 21637807 (PMID 21637807)
Citation will be completed automatically in a few minutes. Jump the queue or expand by hand
External links
- Pages with script errors
- CS1 maint: Multiple names: authors list
- Pages with incomplete PMID references
- Articles without InChI source
- Chemical pages without ChemSpiderID
- Articles without KEGG source
- Articles without UNII source
- Chemical articles with unknown parameter in Chembox
- Articles with changed EBI identifier
- ECHA InfoCard ID from Wikidata
- Articles containing unverified chemical infoboxes
- Chembox image size set
- Pages with broken file links
- All articles with links needing disambiguation
- Articles with links needing disambiguation from May 2013
- Articles with invalid date parameter in template
- B vitamins
- Phosphates