Candida albicans
| Candida albicans | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||||
| Scientific classification | ||||||||||||||||
| ||||||||||||||||
| Binomial name | ||||||||||||||||
| Candida albicans (C.P. Robin) Berkhout 1923 | ||||||||||||||||
| Synonyms | ||||||||||||||||
|
Candidiasis Main page |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Candida albicans is a diploid fungus (a form of yeast), which is capable of mating but not of meiosis, and a causal agent of opportunistic oral and genital infections in humans.[3][4] Systemic fungal infections (fungemias) have emerged as important causes of morbidity and mortality in immunocompromised patients (e.g., AIDS, cancer chemotherapy, organ or bone marrow transplantation). In addition, hospital-related infections in patients not previously considered at risk (e.g. patients on an intensive care unit) have become a cause of major health concern.
C. albicans is among the gut flora, the many organisms which live in the human mouth and gastrointestinal tract. Under normal circumstances, C. albicans lives in 80% of the human population with no harmful effects, although overgrowth results in candidiasis. Candidiasis is often observed in immunocompromised individuals such as HIV-positive patients. Candidiasis also may occur in the blood and in the genital tract. Candidiasis, also known as "thrush", is a common condition which is usually easily cured in people who are not immunocompromised. To infect host tissue, the usual unicellular yeast-like form of Candida albicans reacts to environmental cues and switches into an invasive, multicellular filamentous form.[3]
Genome
One of the most interesting features of the C. albicans genome is the occurrence of numeric and structural chromosomal rearrangements as means of generating genetic diversity, named chromosome length polymorphisms (contraction/expansion of repeats), reciprocal translocations, chromosome deletions and trisomy of individual chromosomes. These karyotypic alterations lead to changes in the phenotype, which is an adaptation strategy of this fungus. These mechanisms will be better understood with the complete analysis of the C. albicans genome.
The Candida albicans genome for strain SC5314 was sequenced at the Stanford DNA Sequencing and Technology Center.[5][6] The genome of the WO1 strain was sequenced by the Broad Institute of MIT and Harvard.[7]
The sequencing of the C. albicans genome and subsequently of the genomes of several other medically relevant Candida species has profoundly and irreversibly changed the way Candida species are now investigated and understood.[4] The C. albicans genome sequencing effort was launched in October 1996. Successive releases of the sequencing data and genome assemblies have marked the last 10 years, culminating with the release of the diploid assembly 19 which provided a haploid version of the genome along with data on allelic regions in the genome.[4] A refined assembly 20 with the eight assembled C. albicans chromosomes was released in the summer of 2006. Importantly, the availability of sequencing data prior to the completion of the genome sequence has made it possible to start C. albicans post-genomics early on. In this regard, genome databases have been made available to the research community providing different forms of genome annotation. These have been merged in a community-based annotation hosted by the Candida Genome Database. The availability of the genome sequence has paved the way for the implementation of post-genomic approaches to the study of C. albicans: macroarrays and then microarrays have been developed and used to study the C. albicans transcriptome; proteomics has also been developed and complements transcriptional analyses; furthermore, systematic approaches are becoming available to study the contribution of each C. albicans gene in different contexts. Other Candida genome sequences have been, or are being, determined: C. glabrata, C. dubliniensis, C. parapsilosis, C. guilliermondii, C. lusitaniae, and C. tropicalis. These species will soon enter the post-genomic era as well and provide interesting comparative data. The genome sequences obtained for the different Candida species along with those of non-pathogenic hemiascomycetes provide a wealth of knowledge on the evolutionary processes which have shaped the hemiascomycete group as well as those which may have contributed to the success of different Candida species as pathogens.[4]
The genome of C. albicans is highly dynamic and this variability has been used advantageously for molecular epidemiological studies of C. albicans and population studies in this species. A remarkable discovery which has arisen from the genome sequence is the presence of a parasexual cycle in C. albicans. This parasexual cycle is under the control of mating-type loci and switching between white and opaque phenotypes. Investigating the role which the mating process plays in the dynamics of the C. albicans population or in other aspects of C. albicans biology and pathogenicity will undoubtedly represent an important focus for future research.[4]
Dimorphism
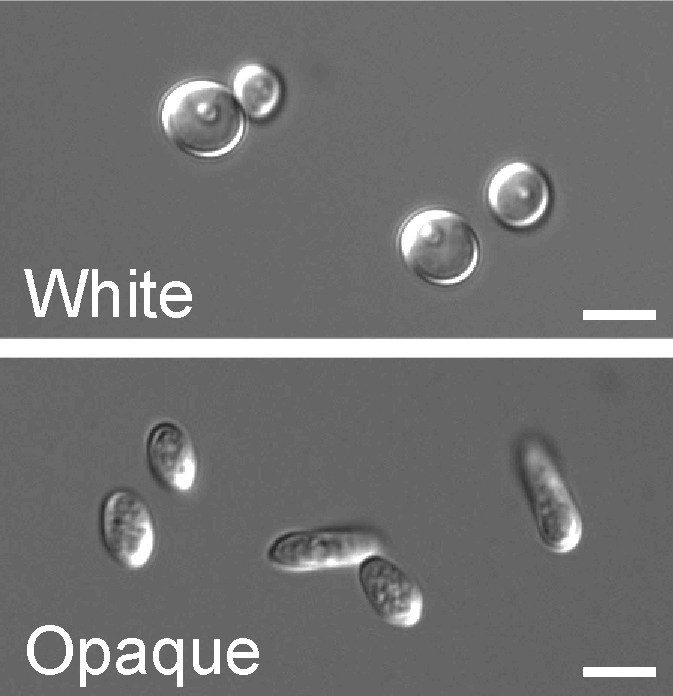
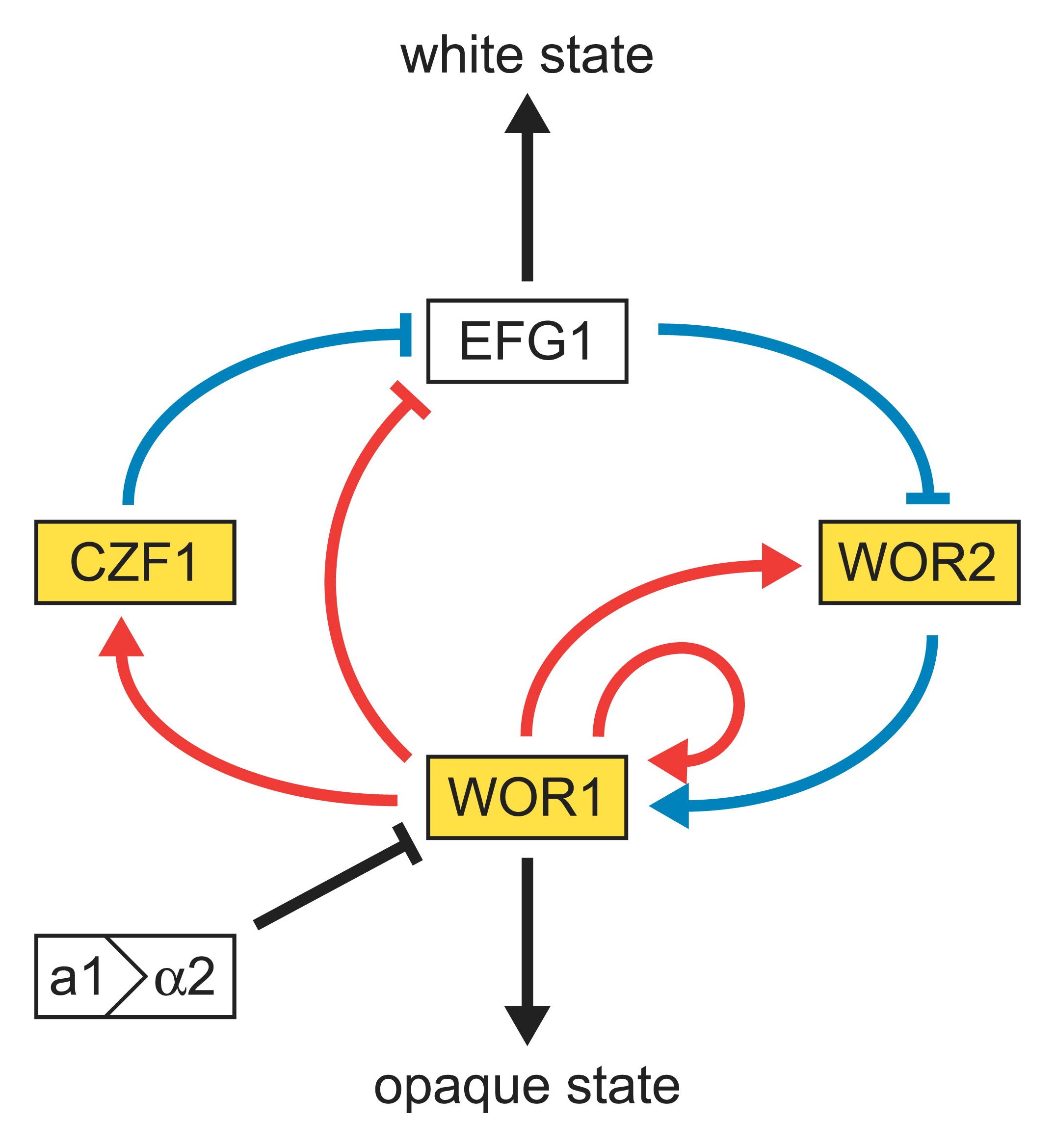
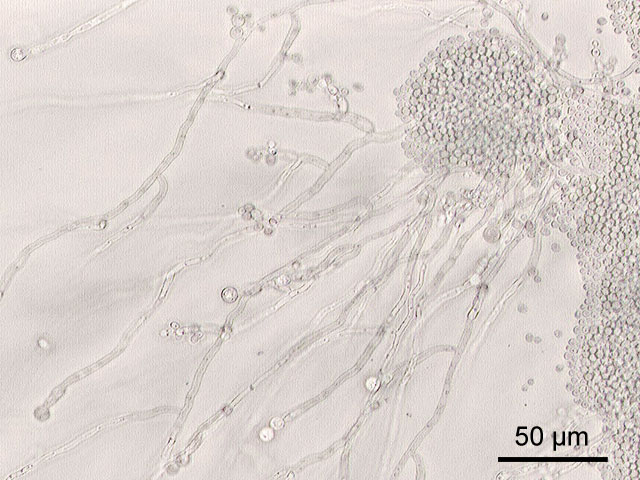
In a process which superficially resembles dimorphism, C. albicans undergoes a process called phenotypic switching, in which different cellular morphologies are generated spontaneously. One of the classically studied strains which undergoes phenotypic switching is WO-1, which consists of two phases - one which grows as smooth white colonies and one which is rod-like and grows as flat gray colonies. The other strain known to undergo switching is 3153A; this strain produces at least seven different colony morphologies. In both the WO-1 and 3153A strains, the different phases convert spontaneously to the other(s) at a low frequency. The switching is reversible, and colony type can be inherited from one generation to another. While several genes which are expressed differently in different colony morphologies have been identified, some recent efforts have focused on what might be controlling these changes. Further, whether there is a potential molecular link between dimorphism and phenotypic switching is a tantalizing question.
In the 3153A strain, a gene called SIR2 (for silent information regulator) has been found which seems to be important for phenotypic switching. SIR2 was originally found in Saccharomyces cerevisiae (brewer's yeast), where it is involved in chromosomal silencing — a form of transcriptional regulation in which regions of the genome are reversibly inactivated by changes in chromatin structure (chromatin is the complex of DNA and proteins which make chromosomes). In yeast, genes involved in the control of mating type are found in these silent regions, and SIR2 represses their expression by maintaining a silent-competent chromatin structure in this region. The discovery of a C. albicans SIR2 which is implicated in phenotypic switching suggests that it too has silent regions controlled by SIR2, in which the phenotype-specific genes may perhaps reside.
Another potential regulatory molecule is Efg1p, a transcription factor found in the WO-1 strain which regulates dimorphism, and more recently has been suggested to help regulate phenotypic switching. Efg1p is expressed only in the white and not in the gray cell-type, and overexpression of Efg1p in the gray form causes a rapid conversion to the white form.
So far there are few data which say that dimorphism and phenotypic switching use common molecular components. However, it is not inconceivable that phenotypic switching may occur in response to some change in the environment as well as being a spontaneous event. How SIR2 itself is regulated in Saccharomyces cerevisiae may yet provide clues as to the switching mechanisms of C. albicans.
Heterozygosity
The heterozygosity of the Candida genome exceeds that found in other genomes and is widespread among clinical isolates. Non-synonymous single base polymorphisms result in two proteins that differ in one or several amino acids that may confer functional differences for each protein. This situation considerably increases the number of different proteins encoded by the genome.[8]
Gallery
-
photomicrograph of the fungus Candida albicans. From Public Health Image Library (PHIL). [9]
-
Histopathologic changes associated with a fungal infection, which had spread to this rabbit kidney tissue sample, due to the pathogen Candida albicans.(125x mag). From Public Health Image Library (PHIL). [9]
-
Wet mounted vaginal smear specimen, revealed the presence of Candida albicans, which had been extracted from a patient with vaginal candidiasis. From Public Health Image Library (PHIL). [9]
-
Candida albicans fungal organisms in their yeast stage of development (1200x mag). From Public Health Image Library (PHIL). [9]
-
Female perineum highlights the inflammatory reaction known as vulvitis, or vulvovaginal candidiasis, which in this case, was caused by an infection by the fungal organism, Candida albicans. From Public Health Image Library (PHIL). [9]
-
Sputum specimen reveals the presence of chlamydospores of the fungal organism, Candida albicans, in a case of invasive pulmonary candidiasis. From Public Health Image Library (PHIL). [9]
-
Plate culture of the fungus Candida albicans. From Public Health Image Library (PHIL). [9]
See also
- Torula yeast (Candida utilis)
- Psoriasis
- Undecylenic acid (Castor oil derivative) for candida fungus infections.
References
- ↑ Candida albicans at NCBI Taxonomy browser, url accessed 2006-12-26
- ↑ "Factors Affecting the Morphology of Candida Albicans" Dan Otho McClary Annals of the Missouri Botanical Garden, Vol. 39, No. 2 (May, 1952), pp. 137-164. doi:10.2307/2394509
- ↑ Jump up to: 3.0 3.1 Ryan KJ; Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed. ed.). McGraw Hill. ISBN 0838585299.
- ↑ Jump up to: 4.0 4.1 4.2 4.3 4.4 dEnfert C; Hube B (editors) (2007). Candida: Comparative and Functional Genomics. Caister Academic Press. ISBN 9781904455134.
- ↑ Jones T, Federspiel N, Chibana H, Dungan J, Kalman S, Magee B, Newport G, Thorstenson Y, Agabian N, Magee P, Davis R, Scherer S (2004). "The diploid genome sequence of Candida albicans". Proc Natl Acad Sci U S A. 101 (19): 7329–34. PMID 15123810.
- ↑ Braun B, van Het Hoog M, d'Enfert C, Martchenko M, Dungan J, Kuo A, Inglis D, Uhl M, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, Bachewich C, Harcus D, Marcil A, Dignard D, Iouk T, Zito R, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro C, Bates S, Gow N, Hoyer L, K�hler G, Morschh�user J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Costanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell A, Johnson A, Whiteway M, Nantel A (2005). "A human-curated annotation of the Candida albicans genome". PLoS Genet. 1 (1): 36–57. PMID 16103911. replacement character in
|author=at position 282 (help) - ↑ http://www.broad.mit.edu/annotation/genome/candida_albicans/
- ↑ Larriba G; Calderone RA (2008). "Heterozygosity and Loss of Heterozygosity in Candida albicans". Pathogenic Fungi: Insights in Molecular Biology. Caister Academic Press. ISBN 978-1-904455-32-5.
- ↑ Jump up to: 9.0 9.1 9.2 9.3 9.4 9.5 9.6 "Public Health Image Library (PHIL)".
External links
- U.S. National Institutes of Health on the Candida albicans genome
- Causes for candida albicans & Natural Herbal Treatment, FAQ
- NIH - How Candida albicans switches phenotype
- Candida albicans genome
- Candida genomics
- Eczema, psoriasis, Chronic Rashes, and their relationship to Candida albicans
- Incidence of Candida in psoriasis--a study on the fungal flora of psoriatic patients
![photomicrograph of the fungus Candida albicans. From Public Health Image Library (PHIL). [9]](/images/a/ae/Candidiasis_16.jpeg)
![Histopathologic changes associated with a fungal infection, which had spread to this rabbit kidney tissue sample, due to the pathogen Candida albicans.(125x mag). From Public Health Image Library (PHIL). [9]](/images/7/7e/Candidiasis_07.jpeg)
![Wet mounted vaginal smear specimen, revealed the presence of Candida albicans, which had been extracted from a patient with vaginal candidiasis. From Public Health Image Library (PHIL). [9]](/images/7/7a/Candidiasis_06.jpeg)
![Candida albicans fungal organisms in their yeast stage of development (1200x mag). From Public Health Image Library (PHIL). [9]](/images/6/6e/Candidiasis_03.jpeg)
![Female perineum highlights the inflammatory reaction known as vulvitis, or vulvovaginal candidiasis, which in this case, was caused by an infection by the fungal organism, Candida albicans. From Public Health Image Library (PHIL). [9]](/images/3/33/Candidiasis_02.jpeg)
![Sputum specimen reveals the presence of chlamydospores of the fungal organism, Candida albicans, in a case of invasive pulmonary candidiasis. From Public Health Image Library (PHIL). [9]](/images/5/56/Candidiasis_01.jpeg)
![Plate culture of the fungus Candida albicans. From Public Health Image Library (PHIL). [9]](/images/1/15/Candidiasis_19.jpeg)