Dominance relationship
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
In genetics, a dominance relationship refers to how the alleles for a locus interact to produce a phenotype.
There are three main kinds of dominance relationships:
- Simple dominance or complete dominance (simple Mendelian inheritance) over a recessive trait
- Incomplete dominance
- Codominance
In general terms, a gene contributes to production or expression of some trait or character (a phene), e.g. the color of one's iris. A locus is similar, but more loosely defined: since phenes are usually readily apparent but determining which genes contribute to them is a complicated process, loci are simply locations in the genome which are known to be directly related to expression of defined phenes. Most loci can eventually be assigned to specific genes, but there is no straightforward 1:1 relationship between loci and genes, and whereas a gene always encodes for a protein, a locus might be in a noncoding DNA sequence.
An allele (or allelic variant) is any of the versions of some genetic locus that might exist in a population. Regarding the iris example, there exists an allele of the locus called EYCL3 which causes the eye to be some shade of brown from lots of eumelanins, and another that causes few eumelanin to be produced, resulting in an iris that is blue, gray or green. Since you are diploid, you have two copies of the locus, one inherited from your father and the other from your mother. Thus, you can be homozygous in that locus - having one of the two allelic variants twice - or heterozygous, with one copy of each variant.
Brown eye color is the dominant trait in the iris example, whereas non-brown eye color is the recessive trait; often, non-brown is called simply "blue" due to the rarity of green and grey eyes, but this is technically incorrect (see next paragraph). If one or both of your EYCL3 loci carry the allele for the dominant trait, this trait - brown eyes - will be expressed. The "brown eyes" allele induces copious eumelanin production in the iris, whereas the "non-brown" allele causes the production of only small amounts of eumelanins. Therefore, the recessive trait in this example is simply overwhelmed by the dominant trait.
To have green, blue or grey eyes, both copies of the EYCL3 locus must carry the recessive allele, to prevent strong eumelanin production in the iris. In these cases, your ultimate eye color depends on the alleles present at 2 other loci, EYCL1 and EYCL2. Green eyes are dominant over blue eyes, and these alleles are carried at the EYCL1 locus. Here, the pigment expressed is a yellow one, which combined with the bluish hue of a bit of eumelanin gives a green eye color. Persons whose eyes produce large amounts of both pigments - brown and yellow - will have amber eyes. This, incidentially, shows that just because an allele produces a dominant trait, it is not necessarily common: green and amber eyes, although the more dominant trait, are actually rarer than blue eyes. Simply put, the frequency of an allele is due to population genetics effects, whereas the dominant vs recessiveness of a trait is due to how specific biochemical reactions are affected by the different alleles.
Note that it is phenes and phenotypes that are dominant and recessive, not the alleles or genes. Note also that the term "dominant/recessive allele", while technically incorrect, is correct as regards the allele - locus/gene relationship in dominant-recessive inheritance. "Dominant/recessive gene" is utterly wrong in most of the (Mendelian) contexts it is usually encountered; for a gene to dominate over another in expression of a phenotype, epistatic or other forms of multi-gene expressions are required.
Nomenclature
Loci are indicated in shorthand by a combination of one or a few letters - for example, in cat coat genetics the alleles Mc and mc (for "mackerel tabby") play a prominent role. Alleles producing dominant traits are denoted by initial capital letters; those that confer recessive traits are written with lowercase letters. The alleles present in a locus are usually separated by a slash; in the Mc - mc case, the dominant trait is the "mackerel-stripe" pattern, and the recessive one the "classic" or "oyster" tabby pattern, and thus a classical-pattern tabby cat would carry the alleles mc/mc, whereas a mackerel-stripe tabby would be either Mc/mc or Mc/Mc.
For another example, flower color in sweet peas (Lathyrus odoratus) is controlled by a single gene with two alleles. The three genotypes are P/P, P/p, and p/p. The flower color for P/P (purple) and p/p (white) do not depend on the dominance relationship. However, the heterozygote Pp could theoretically have many different colors, e.g., purple, white, or a light purple. The exact color of flowers produced by the heterozygous plants depends on factors other than the dominance relationship between the two alleles in question.
Relationship to other genetics concepts
Humans have 23 homologous chromosome pairs (22 pairs of autosomal chromosomes and two distinct sex chromosomes, X and Y). It is estimated that the human genome contains 20,000-25,000 genes "[2]". Each chromosomal pair has the same genes, although it is generally unlikely that homologous genes from each parent will be identical in sequence. The specific variations possible for a single gene are called alleles: for a single eye-color gene, there may be a blue eye allele, a brown eye allele, a green eye allele, etc. Consequently, a child may inherit a blue eye allele from their mother and a brown eye allele from their father. The dominance relationships between the alleles control which traits are and are not expressed.
An example of an autosomal dominant human disorder is Huntington's disease, which is a neurological disorder resulting in impaired motor function. The mutant gene results in an abnormal protein, containing large repeats of the amino acid glutamine. This defective protein is toxic to neural tissue, resulting in the characteristic symptoms of the disease.
A list of human traits that follow a simple inheritance pattern can be found in human genetics. Humans have several genetic diseases, often but not always caused by recessive genes.
Punnett square
The genetic combinations possible with simple dominance can be expressed by a diagram called a Punnett square. One parent's alleles are listed across the top and the other parent's alleles are listed down the left side. The interior squares represent possible offspring, in the ratio of their statistical probability. In the previous example of flower color, P represents the dominant purple-colored allele and p the recessive white-colored allele. If both parents are purple-colored and heterozygous (Pp), the Punnett square for their offspring would be:
| P | p | |
| P | P P | P p |
| p | P p | p p |
In the PP and Pp cases, the offspring is purple colored due to the dominant P. Only in the pp case is there expression of the recessive white-colored phenotype.
Dominant allele
Dominant trait refers to a genetic feature that hides the recessive trait in the phenotype of an individual. A dominant trait causes the phenotype that is seen in a heterozygous (Aa) genotype. Many traits are determined by pairs of complementary genes, each inherited from a single parent. Often when these are paired and compared, one allele (the dominant) will be found to effectively shut out the instructions from the other, recessive allele. For example, if a person has one allele for blood type A and one for blood type O, that person will always have blood type A because it is the dominant allele. For a person to have blood type O, both their alleles must be O (recessive). When a person has two dominant alleles, they are referred to as homozygous dominant. If they have one dominant allele and one recessive allele, they are referred to as heterozygous.
A dominant trait when written in a genotype is always written before the recessive gene in a heterozygous pair. A heterozygous genotype is written Aa, not aA.
Types of dominances
Simple dominance or complete dominance
Consider the simple example in peas of flower color, first studied by Gregor Mendel. The dominant allele is purple and the recessive allele is white.[verification needed] In a given individual, the two corresponding alleles of the chromosome pair fall into one of three patterns:
- both alleles purple (PP)
- both alleles white (pp)
- one allele purple and one allele white (Pp)
If the two alleles are the same (homozygous), the trait they represent will be expressed. But if the individual carries one of each allele (heterozygous), only the dominant one will be expressed. The recessive allele will simply be suppressed.
Simple dominance in pedigrees
Dominant traits are recognizable by the fact that they do not skip generations, as recessive traits do. It is therefore quite possible for two parents with purple flowers to have a white flowers among their progeny, but two such white offspring could not have purple offspring (although very rarely, one might be produced by mutation). In this situation, the purple individuals in the first generation must have both been heterozygous (carrying one copy of each allele).
Incomplete dominance
Discovered by Karl Correns, incomplete dominance (sometimes called partial dominance) is a heterozygous genotype that creates an intermediate phenotype. In this case, only one allele (usually the wild type) at the single locus is expressed, and the expression is doseage dependent. Two copies of the gene produce full expression, while one copy of the gene produces partial expression in an intermediate phenotype. A cross of two intermediate phenotypes (= monohybrid heterozygotes) will result in the reappearance of both parent phenotypes and the intermediate phenotype. There is a 1:2:1 phenotype ratio instead of the 3:1 phenotype ratio found when one allele is dominant and the other is recessive. This lets an organism's genotype can be diagnosed from its phenotype without time-consuming breeding tests.
The classic example of this is the colors of carnations.
| R | R' | |
| R | RR | RR' |
| R' | RR' | R'R' |
R is the allele for red pigment. R' is the allele for no pigment.
Thus, RR offspring make a lot of red pigment and appear red. R'R' offspring make no red pigment and appear white. Both RR' and R'R offspring make some pigment and therefore appear pink.
A readily visible example of incomplete dominance is the color modifier Merle in dogs.
Codominance
In codominance, neither phenotype is completely dominant. Instead, the heterozygous individual expresses both phenotypes. A common example is the ABO blood group system. The gene for blood types has three alleles: A, B, and i. i causes O type and is recessive to both A and B. The A and B alleles are codominant with each other. When a person has both an A and a B allele, the person has type AB blood.
When two persons with AB blood type have children, the children can be type A, type B, or type AB. There is a 1A:2AB:1B phenotype ratio instead of the 3:1 phenotype ratio found when one allele is dominant and the other is recessive. This is the same phenotype ratio found in matings of two organisms that are heterozygous for incomplete dominant alleles.
Example Punnett square for a father with A and i, and a mother with B and i:
| A | i | |
| B | AB | B |
| i | A | O |
Amongst the very few codominant genetic diseases in humans, one relatively common one is A1AD, in which the genotypes Pi00, PiZ0, PiZZ, and PiSZ all have their more-or-less characteristic clinical representations.
Most molecular markers are considered to be codominant.
A roan horse has codominant follicle genes, expressing individual red and white follicles.
Dominant negative
Most loss of-function mutations are recessive. However, some are dominant and are called "dominant negative" or antimorphic mutations. Typically, a dominant negative mutation occurs when the gene product adversely affects the normal, wild-type gene product within the same cell. This usually occurs if the product can still interact with the same elements as the wild-type product, but block some aspect of its function. Such proteins may be competitive inhibitors of the normal protein functions.
Types:
- A mutation in a transcription factor that removes the activation domain, but still contains the DNA binding domain. This product can then block the wild-type transcription factor from binding the DNA site leading to reduced levels of gene activation.
- A protein that is functional as a dimer. A mutation that removes the functional domain, but retains the dimerization domain would cause a dominate negative phenotype, because some fraction of protein dimers would be missing one of the functional domains.
Autosomal dominant gene
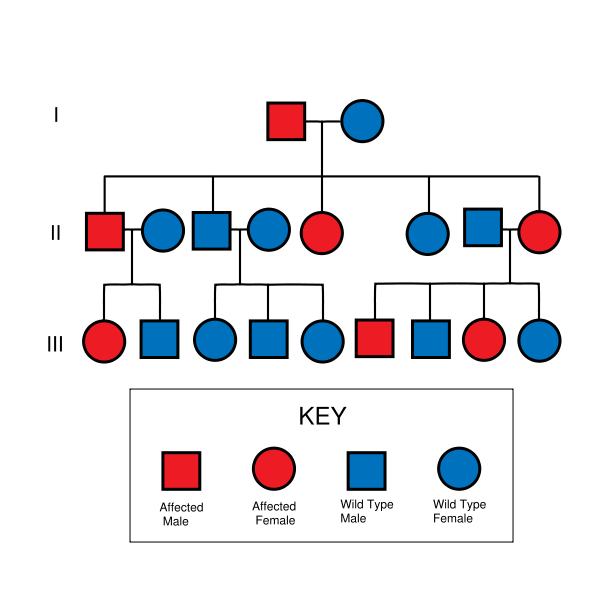
An autosomal dominant gene is one that occurs on an autosomal (non-sex determining) chromosome. As it is dominant, the phenotype it gives will be expressed even if the gene is heterozygous. This contrasts with recessive genes, which need to be homozygous to be expressed.
The chances of an autosomal dominant disorder being inherited are 50% if one parent is heterozygous for the mutant gene and the other is homozygous for the normal, or 'wild-type', gene. This is because the offspring will always inherit a normal gene from the parent carrying the wild-type genes, and will have a 50% chance of inheriting the mutant gene from the other parent. If the mutant gene is inherited, the offspring will be heterozygous for the mutant gene, and will suffer from the disorder. If the parent with the disorder is homozygous for the gene, the offspring produced from mating with an unaffected parent will always have the disorder. See Mendelian inheritance.
The term vertical transmission refers to the concept that autosomal dominant disorders are inherited through generations. This is obvious when you examine the pedigree chart of a family for a particular trait. Because males and females are equally affected, they are equally likely to have affected children.
Although the mutated gene should be present in successive generations in which there are more than one or two offspring, it may appear that a generation is skipped if there is reduced penetrance.
Examples
Autosomal dominant disorders
- Achondroplasia
- Alexander disease
- Antithrombin deficiency
- BRCA1 and BRCA2 mutations (Hereditary Breast Ovarian Cancer Syndrome)
- Brugada syndrome
- Charcot-Marie-Tooth Syndrome
- Ectrodactyly
- Cleft Chin
- Ehlers-Danlos Syndrome
- Familial hypercholesterolemia
- Facioscapulohumeral muscular dystrophy
- Fatal Familial Insomnia
- Fibrodysplasia Ossificans Progressiva
- FOXP2 Gene
- Hereditary hemorrhagic telangiectasia (Osler-Weber-Rendu Syndrome)
- Hereditary multiple exostoses
- Hereditary spherocytosis
- Huntington's Disease
- Hypertrophic cardiomyopathy
- Kabuki syndrome (potentially)
- Lactase Persistence
- Long QT Syndrome
- Malignant hyperthermia
- Mandibulofacial dysostosis (Treacher Collins syndrome)
- Marfan Syndrome
- Mowat-Wilson Syndrome
- Multiple endocrine neoplasia
- Neurofibromatosis
- Osteogenesis Imperfecta
- Pfeiffer syndrome
- Autosomal Dominant Polycystic Kidney Disease, ADPKD (Adult-onset)
- Tourette's Syndrome
- Tuberous Sclerosis
- Von Hippel-Lindau disease
- Familial adenomatous polyposis
- Noonan Syndrome
Recessive allele
The term "recessive gene" refers to an allele that causes a phenotype (visible or detectable characteristic) that is only seen in homozygous genotype (an organism that has two copies of the same allele) and never in a heterozygous genotype. Every person has two copies of every gene on autosomal chromosomes, one from mother and one from father. If a genetic trait is recessive, a person needs to inherit two copies of the gene for the trait to be expressed. Thus, both parents have to be carriers of a recessive trait in order for a child to express that trait. If both parents are carriers, there is a 25% chance with each child to show the recessive trait.
The term "recessive gene" is part of the laws of Mendelian inheritance created by Gregor Mendel. Examples of recessive genes in Mendel's famous pea plant experiments include those that determine the color and shape of seed pods, and plant height.
Autosomal recessive gene
Autosomal recessive is a mode of inheritance of genetic traits located on the autosomes (the 22 non-sex determining chromosomes).
In opposition to autosomal dominant trait, a recessive trait only becomes phenotypically apparent when two copies of a gene (two alleles) are present. In other words, the subject is homozygous for the trait.
Recessive genes will also show a horizontal inheritance on a pedigree chart.
The frequency of the carrier state can be calculated by the Hardy-Weinberg formula: <math> p^2+2pq+q^2=1 </math> (p is the frequency of one pair of alleles, and q = 1 − p is the frequency of the other pair of alleles.)
Recessive genetic disorders occur when both parents are carriers and each contributes an allele to the embryo, meaning these are not dominant genes. As both parents are heterozygous for the disorder, the chance of two disease alleles landing in one of their offspring is 25% (in autosomal dominant traits this is higher). 50% of the children (or 2/3 of the remaining ones) are carriers. When one of the parents is homozygous, the trait will only show in his/her offspring if the other parent is also a carrier. In that case, the chance of disease in the offspring is 50%.
Nomenclature of recessiveness
Technically, the term "recessive gene" is imprecise because it is not the gene that is recessive but the phenotype (or trait). It should also be noted that the concepts of recessiveness and dominance were developed before a molecular understanding of DNA and before molecular biology, thus mapping many newer concepts to "dominant" or "recessive" phenotypes is problematic. Many traits previously thought to be recessive have mild forms or biochemical abnormalities that arise from the presence of the one copy of the allele. This suggests that the dominant phenotype is dependent upon having two dominant genes and the presence of one dominant and one recessive gene creates some blending of both dominant and recessive traits.
Examples
Pea Plant
Gregor Mendel performed many experiments on pea plant (Pisum sativum) while researching traits, chosen because of the simple and low variety of characteristics, as well as the short period of germination. He experimented with color (green vs. yellow), size (short vs. tall), pea texture (smooth vs. wrinkled), and many others. By good fortune, the characteristics displayed by these plants clearly exhibited a dominant and recessive form. This is not true for many organisms.
For example, when testing the color of the pea plants, he chose two yellow plants, since yellow was more common than green. He mated them, and examined the offspring. He continued to mate only those that appeared yellow, and eventually, the green ones would stop being produced. He also mated the green ones together and determined that only green ones were produced.
Mendel determined that this was because green was a recessive trait which only appeared when yellow, the dominant trait, was not present. Also, he determined that the dominant trait would be displayed whether or not the recessive trait was there.
Autosomal recessive disorders
Dominance/recessiveness refers to phenotype, not genotype. An example to prove the point is sickle cell anemia. The sickle cell genotype is caused by a single base pair change in the beta-globin gene: normal=GAG (glu), sickle=GTG (val). There are several phenotypes associated with the sickle genotype:-
- anemia (a recessive trait)
- blood cell sickling (co-dominant)
- altered beta-globin electrophoretic mobility (co-dominant)
- resistance to malaria (dominant)
This example demonstrates that one can only refer to dominance/recessiveness with respect to individual phenotypes.
Other recessive disorders: -
- Albinism
- Alpha 1-antitrypsin deficiency
- Bloom's Syndrome
- Certain forms of Spinal muscular atrophy
- Chronic granulomatous disease, p22-, p47- and p67-deficiency CGD is autosomal recessive while gp91PHOX-deficiency CGD is X-linked.
- Congenital adrenal hyperplasia
- Cystic fibrosis
- Dry (otherwise known as "rice-bran") earwax
- Dubin-Johnson syndrome
- Familial Mediterranean fever
- Fanconi anemia
- Friedreich's ataxia
- Galactosemia
- Glucose-6-phosphate dehydrogenase deficiency
- Glycogen storage diseases (most types)
- Haemochromatosis types 1-3
- Homocystinuria
- Mucopolysaccharidoses (most types)
- Pendred syndrome
- Phenylketonuria
- Autosomal Recessive Polycystic Kidney Disease, ARPKD (Child-onset)
- Rotor syndrome
- Tay-Sachs disease
- Thalassemia
- Wilson's disease
- Xeroderma pigmentosum
Mechanisms of dominance
Many genes code for enzymes. Consider the case where someone is homozygous for some trait. Both alleles code for the same enzyme, which causes a trait. Only a small amount of that enzyme may be necessary for a given phenotype. The individual therefore has a surplus of the necessary enzyme. Let's call this case "normal". Individuals without any functional copies cannot produce the enzyme at all, and their phenotype reflects that. Consider a heterozygous individual. Since only a small amount of the normal enzyme is needed, there is still enough enzyme to show the phenotype. This is why some alleles are dominant over others.
In the case of incomplete dominance, the single dominant allele does not produce enough enzyme, so the heterozygotes show some different phenotype. For example, fruit color in eggplants is inherited in this manner. A purple color is caused by two functional copies of the enzyme, with a white color resulting from two non-functional copies. With only one functional copy, there is not enough purple pigment, and the color of the fruit is a lighter shade, called violet.
Some non-normal alleles can be dominant. The mechanisms for this are varied, but one simple example is when the functional enzyme <math> E </math> is composed of several subunits <math>E= E_1\cdots E_n </math> where each <math> E_i </math> is made of several alleles <math> E_i =a_{i1}a_{i2} </math>, with <math> a_{i1},a_{i2}\in\lbrace A_i,A_i'\rbrace </math> making them either functional or not functional according to one of the schemes described above. For example one could have the rule that if any of the <math> E_i </math> subunits are nonfunctional, the entire enzyme <math> E </math> is nonfunctional in the sense that the phenotype is not displayed. In the case of a single subunit say <math> E_1 </math> is <math> E_1 = F </math> where <math> F </math> has a functional and nonfunctional allele (heterozygous individual)(<math> F = a_1A_1 </math>) , the concentration of functional enzyme determined by <math> E </math> could be 50% of normal. If the enzyme has two identical subunits (<math> E = FF\cdots E_n </math> the concentration of functional enzyme is 25% of normal. For four subunits, <math>E=FFFF\cdots E_n </math> the concentration of functional enzyme is about 6% of normal (roughly scaling slower than <math> 1/2^c </math> where <math> c </math> is the number of copies of the allele ( <math> 1/2^4</math> is about 51% percent) This may not be enough to produce the wild type phenotype. There are other mechanisms for dominant mutants.
Other factors
It is important to note that most genetic traits are not simply controlled by a single set of alleles. Often many alleles, each with their own dominance relationships, contribute in varying ways to complex traits.
Some medical conditions may have multiple inheritance patterns, such as in centronuclear myopathy or myotubular myopathy, where the autosomal dominant form is on chromosome 19 but the sex-linked form is on the X chromosome.
See also
References
af:Dominansie (genetika) ca:Codominància de:Dominanz (Genetik) ko:우열의 법칙 it:Eredità autosomica dominante he:יחסי דומיננטיות lv:Kodominance hu:Autoszómális domináns öröklődés sr:Аутозомно-доминантно наслеђивање