Celiac disease pathophysiology
|
Celiac disease Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Medical Therapy |
|
Case Studies |
|
Celiac disease pathophysiology On the Web |
|
American Roentgen Ray Society Images of Celiac disease pathophysiology |
|
Risk calculators and risk factors for Celiac disease pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Celiac disease is caused by a reaction to gliadin, a gluten protein found in wheat (and similar proteins of the tribe Triticeae which includes other cultivars such as barley and rye). Upon exposure to gliadin, the enzyme tissue transglutaminase modifies the protein, and the immune system cross-reacts with the bowel tissue, causing an inflammatory reaction. That leads to flattening of the lining of the small intestine, which interferes with the absorption of nutrients. The only effective treatment is a lifelong gluten-free diet.
Gluten may cause symptoms in people without celiac disease.[1][2]
Pathophysiology
Coeliac disease appears to be polyfactorial, both in that more than one abnormal factor can cause the disease and also more than one factor is necessary for the disease to manifest in a patient.
Most all coeliac patients have abnormal HLA DQ2 allele. However, about 20–30% of people without coeliac disease have inherited an abnormal HLA-DQ2 allele. This suggests additional factors are needed for coeliac disease to develop. Furthermore, about 5% of those people who do develop coeliac disease do not have the DQ2 gene.
The HLA-DQ2 allele shows incomplete penetrance, as the gene alleles associated with the disease appear in most patients, but are neither present in all cases nor sufficient by themselves cause the disease.
Role of other grains
Wheat varieties or subspecies containing gluten such as spelt and Kamut®, and the rye/wheat hybrid triticale, also trigger symptoms.[3]
Barley and rye also induce symptoms of coeliac disease.[3] A small minority of coeliac patients also react to oats.[4][5] Most probably oats produce symptoms due to cross contamination with other grains in the fields or in the distribution channels. There is at least one oat vendor, Gluten Free Oats®, that offers oats that can be considered SAFE for people who are gluten intolerant because they are tested to be below 10 parts per million (ppm) by the University of Nebraska FARRP Laboratory [6]. Another vendor (McCann's) which, while not claiming to be gluten-free, points out that the risk of contamination from their Oats product is low due to the processes they use.[7] Other cereals, such as maize (corn), quinoa, millet, sorghum, rice are safe for a patient to consume. Other carbohydrate-rich foods such as potatoes and bananas do not contain gluten and do not trigger symptoms.
Prolamins
The proteins in food responsible for the immune reaction in coeliac disease are the prolamins. These are storage proteins rich in proline (prol-) and glutamine- (-amin) that dissolve in alcohols and are resistant to pepsin and chymotrypsin, the two main digestive proteases in the gut. Gliadin in wheat is the best-understood member of this family, but other prolamins exist and hordein (from barley), and secalin (from rye) may contribute to coeliac disease. However, not all prolaminins will cause this immune reaction and there is ongoing controversy on the ability of avenin (the prolamin found in oats) to induce this response in coeliac disease.
Tissue transglutaminase
Anti-transglutaminase antibodies to the enzyme tissue transglutaminase (tTG) are found in an overwhelming majority of cases.[8] Tissue transglutaminase modifies gluten peptides into a form that may stimulate the immune system more effectively.
Stored biopsies from suspected coeliac patients has revealed that autoantibody deposits in the subclinical coeliacs are detected prior to clinical disease. These deposits are also found in patients who present with other autoimmune diseases, anemia or malabsorption phenomena at a much increased rate over the normal population.[9] Endomysial component of antibodies (EMA) to tTG are believed to be directed toward cell surface transglutaminase, and these antibodies are still used in confirming a coeliac disease diagnosis. However, a 2006 study showed that EMA-negative coeliac patients tend to be older males with more severe abdominal symptoms and a lower frequency of "atypical" symptoms including autoimmune disease.[10] In this study the anti-tTG antibody deposits did not correlate with the severity of villous destruction. These findings, coupled with recent work showing that gliadin has an innate response component,[11] suggests that gliadin may be more responsible for the primary manifestations of coeliac disease whereas tTG is a bigger factor in secondary effects such as allegic responses and secondary autoimmune diseases. In a large percentage of coeliac patients the anti-tTG antibodies also recognize a rotavirus protein called VP7. These antibodies stimulate monocytes proliferation and rotavirus infection might explain some early steps in the cascade of immune cell proliferation.[12] Indeed, earlier studies of rotavirus damage in the gut showed this causes a villous atrophy.[13] This suggests that viral proteins may take part in the initial flattening and stimulate self-crossreactive anti-VP7 production. Antibodies to VP7 may also slow healing until the gliadin mediated tTG presentation provides a second source of crossreactive antibodies.
Villous atrophy and malabsorption
The inflammatory process, mediated by T cells, leads to disruption of the structure and function of the small bowel's mucous lining, and causes malabsorption as it impairs the body's ability to absorb nutrients, minerals and fat-soluble vitamins A, D, E and K from food. Lactose intolerance may be present due to the decreased bowel surface and reduced production of lactase but typically resolves once the condition is treated.
Alternative causes of this tissue damage have been proposed and involve release of interleukin 15 and activation of the innate immune system by a shorter gluten peptide (p31–43/49). This would trigger killing of enterocytes by lymphocytes in the epithelium. The villous atrophy seen on biopsy may also be due to unrelated causes, such as tropical sprue, giardiasis and radiation enteritis. While positive serology and typical biopsy are highly suggestive of coeliac disease, lack of response to diet may require these alternative diagnoses to be considered.
Risk modifiers
There are various theories as to what determines whether a genetically susceptible individual will go on to develop coeliac disease. Major theories include infection by rotavirus[14] or human intestinal adenovirus.[15] Some research has suggested that smoking is protective against adult onset coeliac disease.[16]
A 2005 prospective and observational study found that timing of the exposure to gluten in childhood was an important risk modifier. People exposed to wheat, barley, or rye before the gut barrier has fully developed (three months after birth) had five times the risk of developing coeliac disease over those exposed at 4 to 6 months. Those exposed later had a slightly increased risk relative to those exposed at 4 to 6 months.[17] However a 2006 study with similar numbers found just the reverse, that early introduction of grains was protective.[18] Breastfeeding may also reduce risk. A meta-analysis indicates that prolonging breastfeeding until the introduction of gluten-containing grains into the diet was associated with a 52% reduced risk of developing coeliac disease in infancy; whether this persists into adulthood is not clear.[19]
Genetics
The vast majority of coeliac patients have one of two types of HLA DQ.[20] This gene is part of the MHC class II antigen-presenting receptor (also called the human leukocyte antigen) system and distinguishes cells between self and non-self for the purposes of the immune system. There are 7 HLA DQ variants (DQ2 and DQ4 through 9). Two of these variants—DQ2 and DQ8—are associated with coeliac disease. The gene is located on the short arm of the sixth chromosome, and as a result of the linkage this locus has been labeled CELIAC1.
Over 95% of coeliac patients have an isoform of DQ2 (encoded by DQA1*05 and DQB1*02 genes) and DQ8 (encoded by the haplotype DQA1*03:DQB1*0302), which is inherited in families. The reason these genes produce an increase in risk of coeliac disease is that the receptors formed by these genes bind to gliadin peptides more tightly than other forms of the antigen-presenting receptor. Therefore, these forms of the receptor are more likely to activate T lymphocytes and initiate the autoimmune process.
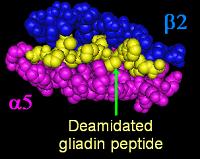
Most coeliac patients bear a two-gene HLA-DQ haplotype referred to as DQ2.5 haplotype. This haplotype is composed of 2 adjacent gene alleles, DQA1*0501 and DQB1*0201, which encode the two subunits, DQ α5 and DQ β2. In most individuals, this DQ2.5 isoform is encoded by one of two chromosomes 6 inherited from parents. Most coeliacs inherit only one copy of this DQ2.5 haplotype, while some inherit it from both parents; the latter are especially at risk for coeliac disease, as well as being more susceptible to severe complications.[22] Some individuals inherit DQ2.5 from one parent and portions of the haplotype (DQB1*02 or DQA1*05) from the other parent, increasing risk. Less commonly, some individuals inherit the DQA1*05 allele from one parent and the DQB1*02 from the other parent, called a trans-haplotype association, and these individuals are at similar risk for coeliac disease as those with a single DQ2.5 bearing chromosome 6, but in this instance disease tends not to be familial. Among the 6% of European celiacs that do not have DQ2.5(cis or trans) or DQ8, 4% are DQ2 and 2% DQA1*05, 0.4% cannot be linked to DQ8, DQA1*05, or DQB1*02.[23]
The frequency of these genes varies geographically. DQ2.5 has high frequency in peoples of North and Western Europe (Basque Country, Ireland,[24] with highest frequencies), portions of Africa, and is associated disease in India,[25] but is not found along portions of the West Pacific rim. DQ8, spread more globally than DQ2.5, is more prevalent from South and Central America (up to 90% phenotype frequency).[26]
In addition to the CELIAC1 locus, CELIAC2 (5q31-q33 - IBD5 locus), CELIAC3 (2q33 -CTLA4 locus), CELIAC4 (19q13.1 - MYOIXB locus), have been linked to coeliac disease. The CTLA4 and myosin IXB and gene have been found to be linked to coeliac disease and other autoimmune diseases.[27][28] Two additional loci on chromosome 4, 4q27 (IL2 or IL21 locus) and 4q14, have been found to be linked to coeliac disease.[29][30]
HLA genetic typing
Antibody testing and HLA testing have similar accuracies[20]
Pathology
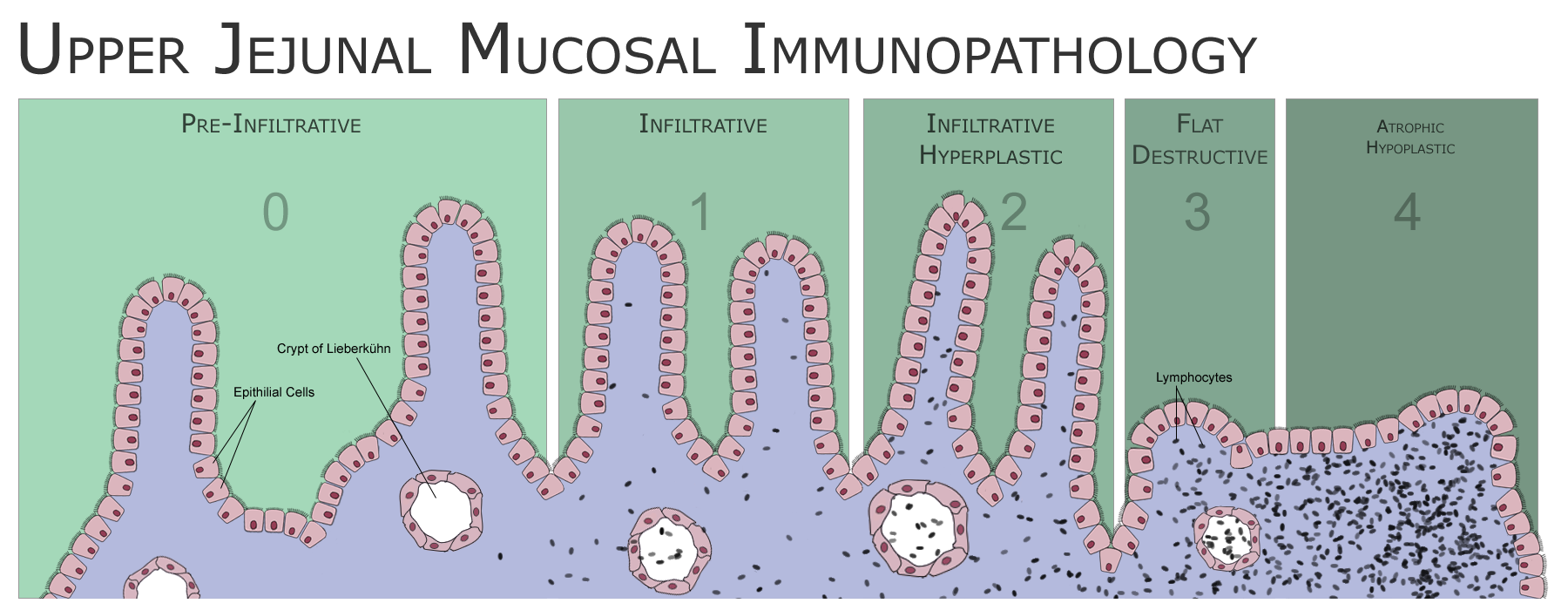
The classic pathology changes of coeliac disease in the small bowel are categorized by the "Marsh classification":[31]
- Marsh stage 0: normal mucosa
- Marsh stage 1: increased number of intra-epithelial lymphocytes, usually exceeding 20 per 100 enterocytes
- Marsh stage 2: proliferation of the crypts of Lieberkuhn
- Marsh stage 3: partial or complete villous atrophy
- Marsh stage 4: hypoplasia of the small bowel architecture
The changes classically improve or reverse after gluten is removed from the diet, so many official guidelines recommend a repeat biopsy several (4–6) months after commencement of gluten exclusion.
In some cases a deliberate gluten challenge, followed by biopsy, may be conducted to confirm or refute the diagnosis. A normal biopsy and normal serology after challenge indicates the diagnosis may have been incorrect. Patients are warned that one does not "outgrow" coeliac disease in the same way as childhood food intolerances.
Associated Conditions
- Insulin dependent diabetes mellitus (IDDM)
- Sjogren’s syndrome
- Thyroid disease
- Dermatitis herpetiformis – extraintestinal manifestation
References
- ↑ Biesiekierski JR, Newnham ED, Irving PM, Barrett JS, Haines M, Doecke JD; et al. (2011). "Gluten causes gastrointestinal symptoms in subjects without celiac disease: a double-blind randomized placebo-controlled trial". Am J Gastroenterol. 106 (3): 508–14. doi:10.1038/ajg.2010.487. PMID 21224837.
- ↑ Carroccio A, Mansueto P, Iacono G, Soresi M, D'Alcamo A, Cavataio F; et al. (2012). "Non-celiac wheat sensitivity diagnosed by double-blind placebo-controlled challenge: exploring a new clinical entity". Am J Gastroenterol. 107 (12): 1898–906. doi:10.1038/ajg.2012.236. PMID 22825366.
- ↑ 3.0 3.1 "Grain toxicity" (RTF). The CELIAC list. Retrieved 2006-08-27.
- ↑ Lundin K, Nilsen E, Scott H, Løberg E, Gjøen A, Bratlie J, Skar V, Mendez E, Løvik A, Kett K (2003). "Oats induced villous atrophy in coeliac disease". Gut. 52 (11): 1649–52. PMID 14570737.
- ↑ Størsrud S, Olsson M, Arvidsson Lenner R, Nilsson L, Nilsson O, Kilander A (2003). "Adult coeliac patients do tolerate large amounts of oats". Eur J Clin Nutr. 57 (1): 163–9. PMID 12548312.
- ↑ http://www.glutenfreeoats.com, http://www.farrp.org/, http://www.farrp.org/analysis.htm
- ↑ "McCann's FAQ". Odlum Group. 2004. Retrieved 2006-11-03.
we reckon that the level of non-oat grains to be less than 0.05%
- ↑ Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken E, Schuppan D (1997). "Identification of tissue transglutaminase as the autoantigen of celiac disease". Nat Med. 3 (7): 797–801. PMID 9212111.
- ↑ Kaukinen K, Peraaho M, Collin P, Partanen J, Woolley N, Kaartinen T, Nuuntinen T, Halttunen T, Maki M, Korponay-Szabo I (2005). "Small-bowel mucosal tranglutaminase 2-specific IgA deposits in coeliac disease without villous atrophy: A Prospective and radmonized clinical study". Scand J Gastroenterology. 40: 564–572. PMID 16036509.
- ↑ Salmi T, Collin P, Korponay-Szabó I, Laurila K, Partanen J, Huhtala H, Király R, Lorand L, Reunala T, Mäki M, Kaukinen K (2006). "Endomysial antibody-negative coeliac disease: clinical characteristics and intestinal autoantibody deposits". Gut. 55 (12): 1746–53. PMID 16571636.
- ↑ Londei M, Ciacci C, Ricciardelli I, Vacca L, Quaratino S, and Maiuri L. (2005). "Gliadin as a stimulator of innate responses in celiac disease". Mol Immunol. 42 (8): 913–918. PMID 15829281.
- ↑ Zanoni G, Navone R, Lunardi C, Tridente G, Bason C, Sivori S, Beri R, Dolcino M, Valletta E, Corrocher R, Puccetti A (2006). "In celiac disease, a subset of autoantibodies against transglutaminase binds toll-like receptor 4 and induces activation of monocytes". PLoS Med. 3 (9): e358. PMID 16984219.
- ↑ Salim A, Phillips A, Farthing M (1990). "Pathogenesis of gut virus infection". Baillieres Clin Gastroenterol. 4 (3): 593–607. PMID 1962725.
- ↑ Stene L, Honeyman M, Hoffenberg E, Haas J, Sokol R, Emery L, Taki I, Norris J, Erlich H, Eisenbarth G, Rewers M (2006). "Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study". Am J Gastroenterol. 101 (10): 2333–40. PMID 17032199.
- ↑ Kagnoff M, Paterson Y, Kumar P, Kasarda D, Carbone F, Unsworth D, Austin R (1987). "Evidence for the role of a human intestinal adenovirus in the pathogenesis of coeliac disease". Gut. 28 (8): 995–1001. PMID 2822550.
- ↑ Suman S, Williams E, Thomas P, Surgenor S, Snook J (2003). "Is the risk of adult coeliac disease causally related to cigarette exposure?". Eur J Gastroenterol Hepatol. 15 (9): 995–1000. PMID 12923372.
- ↑ Norris JM, Barriga K, Hoffenberg EJ, Taki I, Miao D, Haas JE, Emery LM, Sokol RJ, Erlich HA, Eisenbarth GS, Rewers M. (2005). "Risk of celiac disease autoimmunity and timing of gluten introduction in the diet of infants at increased risk of disease". JAMA. 293 (19): 2343–2351. PMID 15900004.
- ↑ Poole J, Barriga K, Leung D, Hoffman M, Eisenbarth G, Rewers M, Norris J (2006). "Timing of initial exposure to cereal grains and the risk of wheat allergy". Pediatrics. 117 (6): 2175–82. PMID 16740862.
- ↑ Akobeng A, Ramanan A, Buchan I, Heller R (2006). "Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies". Arch Dis Child. 91 (1): 39–43. PMID 16287899.
- ↑ 20.0 20.1 Hadithi M, von Blomberg BM, Crusius JB; et al. (2007). "Accuracy of serologic tests and HLA-DQ typing for diagnosing celiac disease". Ann. Intern. Med. 147 (5): 294–302. PMID 17785484.
- ↑ Kim C, Quarsten H, Bergseng E, Khosla C, Sollid L (2004). "Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease". Proc Natl Acad Sci U S A. 101 (12): 4175–9. PMID 15020763.
- ↑ Jores RD, Frau F, Cucca F; et al. (2007). "HLA-DQB1*0201 homozygosis predisposes to severe intestinal damage in celiac disease". Scand. J. Gastroenterol. 42 (1): 48–53. doi:10.1080/00365520600789859. PMID 17190762.
- ↑ Karell K, Louka AS, Moodie SJ; et al. (2003). "HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease". Hum. Immunol. 64 (4): 469–77. PMID 12651074.
- ↑ Michalski J, McCombs C, Arai T, Elston R, Cao T, McCarthy C, Stevens F (1996). "HLA-DR, DQ genotypes of celiac disease patients and healthy subjects from the West of Ireland". Tissue Antigens. 47 (2): 127–33. PMID 8851726.
- ↑ Kaur G, Sarkar N, Bhatnagar S; et al. (2002). "Pediatric celiac disease in India is associated with multiple DR3-DQ2 haplotypes". Hum. Immunol. 63 (8): 677–82. PMID 12121676.
- ↑ Layrisse Z, Guedez Y, Domínguez E, Paz N, Montagnani S, Matos M, Herrera F, Ogando V, Balbas O, Rodríguez-Larralde A (2001). "Extended HLA haplotypes in a Carib Amerindian population: the Yucpa of the Perija Range". Hum Immunol. 62 (9): 992–1000. PMID 11543901.
- ↑ Zhernakova A, Eerligh P, Barrera P; et al. (2005). "CTLA4 is differentially associated with autoimmune diseases in the Dutch population". Hum. Genet. 118 (1): 58–66. doi:10.1007/s00439-005-0006-z. PMID 16025348.
- ↑ Sánchez E, Alizadeh BZ, Valdigem G; et al. (2007). "MYO9B gene polymorphisms are associated with autoimmune diseases in Spanish population". Hum. Immunol. 68 (7): 610–5. doi:10.1016/j.humimm.2007.03.006. PMID 17584584.
- ↑ van Heel DA, Franke L, Hunt KA; et al. (2007). "A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21". Nat Genet. 39 (7): 827–9. doi:10.1038/ng2058. PMID 17558408.
- ↑ Popat S, Bevan S, Braegger C, Busch A, O'Donoghue D, Falth-Magnusson K, Godkin A, Hogberg L, Holmes G, Hosie K, Howdle P, Jenkins H, Jewell D, Johnston S, Kennedy N, Kumar P, Logan R, Love A, Marsh M, Mulder C, Sjoberg K, Stenhammar L, Walker-Smith J, Houlston R (2002). "Genome screening of coeliac disease". J Med Genet. 39 (5): 328–31. PMID 12011149.
- ↑ Marsh MN (1992). "Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity ('celiac sprue')". Gastroenterology. 102 (1): 330–54. PMID 1727768. Unknown parameter
|month=ignored (help)