Sideroblastic anemia pathophysiology: Difference between revisions
Nazia Fuad (talk | contribs) (/* Pathophysiology{{cite journal |vauthors=Fujiwara T, Harigae H |title=Pathophysiology and genetic mutations in congenital sideroblastic anemia |journal=Pediatr Int |volume=55 |issue=6 |pages=675–9 |date=December 2013 |pmid=24003969 |doi=10.1111/ped...) |
Nazia Fuad (talk | contribs) |
||
| (22 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{Infobox_Disease | | |||
Name = {{PAGENAME}} | | |||
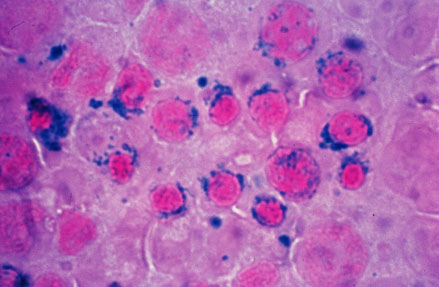
Image = Sideroblastic (microcytic) anemia.jpg | | |||
Caption = Sideroblastic (microcytic) anemia| | |||
}} | |||
{{Sideroblastic anemia}} | {{Sideroblastic anemia}} | ||
{{CMG}} {{AE}} {{N.F}} | {{CMG}} {{AE}} {{N.F}} | ||
| Line 6: | Line 11: | ||
==Overview== | ==Overview== | ||
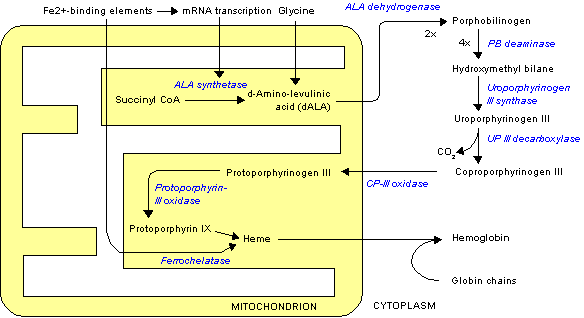
It is understood that sideroblastic anemia is the result of defects in the steps of [[heme]] biosynthesis that occur within the [[Mitochondrion|mitochondrion.]] Sideroblasts are the pathognomic feature of sideroblastic anemia. There is deffect in incorporation of [[iron]] in to [[heme]]. As a result the iron accumulates in mitochondria of red cell precursors. Ring sideroblasts are [[erythroblasts]] that have iron-loaded [[Mitochondrion|mitochondria]]. The [[iron]] granules are arranged around the nucleus in a ring form. They can be seen with prussian blue staining as blue granules around the nucleus.The pathophysiology of sideroblastic anemia depends on the underlying cause. Impaired [[hemoglobin]] production, results in reduced number of mature [[Red blood cell|erythrocytes]]. Resulting [[anemia]] is usually [[Microcytic anemia|microcytic]] and [[Hypochromic anemia|hypochromic]]. The [[iron]] overload in sideroblastic anemia is due to abnormalities in [[iron]] utilization. There is increased iron transport to [[erythroblasts]]. Since the body sense [[anemia]] intestinal iron absorption increases. There is increased iron content in mitochondria of erythroblasts and systemic iron accumulation. Systemic [[iron]] overload occurs only in some forms of sideroblastic anemia, usually when the defects in iron metabolisms involve earlier stages of erythroid pathways. The development of heriditory sideroblastic anemia is the result of multiple [[Mutation|genetic mutations]] in several [[Gene|genes]] involved in heme synthesis resulting in autosomal recessive congenital sideroblastic anemia. Out of many genes ''SLC25A38'' mutations is the most common. | |||
==Pathophysiology== | |||
==Pathophysiology | |||
=== Physiology === | === Physiology === | ||
{| align="right" | |||
|[[File:HemeB.png|400px|thumb|left|Image showing Heme structure[https://commons.wikimedia.org/wiki/File:HemeB.png source:By SubDural12 [Public domain], from Wikimedia Commons]]] | |||
|} | |||
'''Heme''' is porphyrin containing compound, with an Fe [[iron]] ion in the centre,surrounded by [[Heterocyclic compound|heterocyclic]] organic ring of [[porphyrin]]. | |||
The normal physiology of heme synthesis can be understood as follows: | The normal physiology of heme synthesis can be understood as follows:<ref name="pmid20506125">{{cite journal |vauthors=Layer G, Reichelt J, Jahn D, Heinz DW |title=Structure and function of enzymes in heme biosynthesis |journal=Protein Sci. |volume=19 |issue=6 |pages=1137–61 |date=June 2010 |pmid=20506125 |pmc=2895239 |doi=10.1002/pro.405 |url=}}</ref> | ||
* Mitochondria in the developing erythroid cells are the cellular site of heme production and iron utilization. | * [[Mitochondrion|Mitochondria]] in the developing [[Erythroid|erythroid cells]] are the cellular site of [[heme]] production and [[iron]] utilization. | ||
* Glycine combines with succinylcoA to form aminolenolinic acid(ALA) | * [[Glycine]] combines with succinylcoA to form aminolenolinic acid(ALA) | ||
* This reaction is catalyzed by ALA sunthetase enzyme (ALAS2) in mitochondria | * This reaction is catalyzed by ALA sunthetase enzyme (ALAS2) in [[Mitochondrion|mitochondria]] | ||
* ALA synthetase requires vit B-6 as a cofactor | * ALA synthetase requires vit B-6 as a cofactor | ||
* Two molecules of aminolenolinic acid condense in cytosol to form porphobilinogen(PBG) | * Two molecules of aminolenolinic acid [[Condensation|condense]] in [[cytosol]] to form [[porphobilinogen]](PBG) | ||
* This reaction is catalyzed by zinc containing enzyme called ALA dehydratase. | * This reaction is catalyzed by [[zinc]] containing [[enzyme]] called ALA dehydratase. | ||
* Multiple enzymatic transformations in cytoplasm produce coproporphyrinogen III (CPG). | * Multiple enzymatic transformations in [[cytoplasm]] produce [[coproporphyrinogen III]] (CPG). | ||
* Coproporphyrinogen III (CPG) enters the mitochondrion. | * [[Coproporphyrinogen III]] (CPG) enters the [[mitochondrion]]. | ||
* Additional modifications of CPG in mitochondrion produces protoporphyrin IX. | * Additional modifications of CPG in [[mitochondrion]] produces [[protoporphyrin IX]]. | ||
* The final step is the insertion of iron into the protoporphyrin IX ring producing | * The final step is the insertion of iron into the [[protoporphyrin IX]] ring producing [[heme]]. | ||
* This final reaction is catalyzed by enzyme ferrochelatase. | * This final reaction is catalyzed by enzyme [[ferrochelatase]]. | ||
=== Pathogenesis === | === Pathogenesis === | ||
{| align="right" | |||
|[[File:Heme synthesis.png|400px|thumb|left|Image showing Heme synthesis [https://commons.wikimedia.org/wiki/File:Heme_synthesis.png source:By Wmheric [CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0)] [Public domain], from Wikimedia Commons]]] | |||
* It is understood that | |} | ||
* [ | |||
* | * It is understood that sideroblastic anemia is the result of defects in the steps of [[heme]] [[biosynthesis]] that occur within the [[mitochondrion]].<ref name="pmid22160084">{{cite journal |vauthors=Fleming MD |title=Congenital sideroblastic anemias: iron and heme lost in mitochondrial translation |journal=Hematology Am Soc Hematol Educ Program |volume=2011 |issue= |pages=525–31 |date=2011 |pmid=22160084 |doi=10.1182/asheducation-2011.1.525 |url=}}</ref> | ||
* [ | * '''Sideroblasts''' are the pathognomic feature of sideroblastic anemia. | ||
* | ** There is deffect in incorporation of iron in to [[heme]]. | ||
** As a result the iron accumulates in [[Mitochondrion|mitochondria]] of red cell precursors | |||
*** Ring sideroblasts are [[erythroblasts]] that have iron-loaded [[Mitochondrion|mitochondria]]. | |||
*** The iron granules are arranged around the nucleus in a ring form | |||
*** They can be seen with prussian blue staining as blue granules around the [[Cell nucleus|nucleus]].<ref name="pmid21632840">{{cite journal |vauthors=Cazzola M, Invernizzi R |title=Ring sideroblasts and sideroblastic anemias |journal=Haematologica |volume=96 |issue=6 |pages=789–92 |date=June 2011 |pmid=21632840 |pmc=3105636 |doi=10.3324/haematol.2011.044628 |url=}}</ref> | |||
* The [[pathophysiology]] of sideroblastic anemia depends on the underlying cause.<ref name="pmid24003969">{{cite journal |vauthors=Fujiwara T, Harigae H |title=Pathophysiology and genetic mutations in congenital sideroblastic anemia |journal=Pediatr Int |volume=55 |issue=6 |pages=675–9 |date=December 2013 |pmid=24003969 |doi=10.1111/ped.12217 |url=}}</ref> | |||
* The X-linked [[Heredity|hereditar]]<nowiki/>y sideroblastic anemias result from [[Mutation|mutations]] in the [[gene]] encoding [[ALAS2]]. | |||
* [[Isoniazid|Isoniazide]], produces sideroblastic anemia in people who dont use [[pyridoxine]] prophylaxis, | |||
**Pyridoxine is a cofactor for ALAS2 enzyme. | |||
**Its deficiency directly impairs ALAS2 function | |||
* Mitochondrial cytopathies | |||
** Result from deletions of portions of the mitochondrial genome . | |||
** The consequent marked mitochondrial dysfunction causes sideroblastic anemia in these disorders. | |||
* [[Ethanol]] is the most common cause of toxin-induced sideroblastic anemia. | |||
* [[Ethanol]] causes sideroblastic anemia by two mechanisms | |||
** Direct [[Antagonism (chemistry)|antagonism]] to [[pyridoxal phosphate]] | |||
** Dietary deficiency of this compound. | |||
** The [[bone marrow]] shows sideroblasts and [[Vacuole|vacuoles]] in the [[Normoblast|normoblasts]] in [[Alcoholism|ethanol toxicity]]. | |||
* [[Lead]] intoxication causes sideroblastic anemia by inhibiting two [[enzymes]] in [[heme]] pathway: | |||
** ALA dehaydratase | |||
** [[Ferrochelatase]] | |||
* [[Penicillamine]] or triethylene tetramine dihydrochloride (Trientene or TTH) used in treatment of [[Wilson's disease|Wilsons disease]] can produce sideroblastic anemia. | |||
** Excessive chelation produces [[copper deficiency]]. | |||
** [[Copper]] catalyzes the last step in heme [[biosynthesis]], insertion of iron into [[protoporphyrin IX]]. | |||
* Zinc intoxication causes sideroblastic anemia in patients using large amount of [[zinc]] supplements. | |||
* Excessive [[zinc]] reduces serum [[copper]] levels. | |||
'''Mechanism of iron overload''' | |||
* Abnormalities in [[iron]] utilization in sideroblastic anemia | |||
* There is increased [[iron]] transport to [[erythroblasts]] since the body senses [[anemia]]. | |||
* Intestinal [[iron]] absorption increases. | |||
* There is increased [[iron]] content in [[Mitochondrion|mitochondria]] of [[erythroblasts]] and systemic [[iron]] accumulation. | |||
* Systemic [[iron]] overload occurs only in some forms of sideroblastic anemia, usually when the defects in iron metabolisms involve earlier stages of [[Red blood cell|erythroid]] pathways. | |||
== Genetics == | |||
The development of sideroblastic anemia is the result of multiple genetic mutations in several genes involved in heme synthesis resulting in autosomal recessive congenital sideroblastic anemia (ARCSA)<ref name="pmid22160084" /><ref name="pmid24003969" /> | |||
* '''''[[SLC25A38]]''''' –''SLC25A38'' mutations is the most common. ''SLC25A38'' encodes an erythroid-specific mitochondrial amino acid carrier that transports glycine into mitochondria for the first step in heme synthesis. | |||
* '''''[[GLRX5]]''''' – ''GLRX5'' encodes protein used in the synthesis of iron-sulfur (Fe-S) clusters. | |||
* '''''[[HSPA9]]''''' – ''HSPA9'' encodes the mitochondrial HSP70 homologue HSPA9, which is also involved in Fe-S cluster formation. | |||
* '''''FECH''''' – ''FECH'' encodes [[ferrochelatase]], the final [[enzyme]] in the heme synthesis pathway, which inserts an iron atom into [[protoporphyrin IX]] | |||
* '''X-linked sideroblastic anemia with [[ataxia]] (ABCB7 mutation)''' | |||
* '''[[Pearson syndrome]] ( mitochondrial DNA deletion)''' | |||
** [[Pearson syndrome]] is a [[Congenital disorder|congenital]] condition | |||
** It affects multiple systems, | |||
** Results in severe sideroblastic anemia, [[Thrombocytopenia causes|thrombocytopenia]] and [[neutropenia]]. | |||
** [[Pancreatic insufficiency]], [[lactic acidosis]], and [[Delayed milestone|growth retardation]] are other common features. | |||
==References== | ==References== | ||
Latest revision as of 20:08, 19 September 2018
| Sideroblastic anemia pathophysiology | |
 | |
|---|---|
| Sideroblastic (microcytic) anemia |
|
Sideroblastic anemia Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Sideroblastic anemia pathophysiology On the Web |
|
American Roentgen Ray Society Images of Sideroblastic anemia pathophysiology |
|
Risk calculators and risk factors for Sideroblastic anemia pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1] Associate Editor(s)-in-Chief: Nazia Fuad M.D.
Overview
It is understood that sideroblastic anemia is the result of defects in the steps of heme biosynthesis that occur within the mitochondrion. Sideroblasts are the pathognomic feature of sideroblastic anemia. There is deffect in incorporation of iron in to heme. As a result the iron accumulates in mitochondria of red cell precursors. Ring sideroblasts are erythroblasts that have iron-loaded mitochondria. The iron granules are arranged around the nucleus in a ring form. They can be seen with prussian blue staining as blue granules around the nucleus.The pathophysiology of sideroblastic anemia depends on the underlying cause. Impaired hemoglobin production, results in reduced number of mature erythrocytes. Resulting anemia is usually microcytic and hypochromic. The iron overload in sideroblastic anemia is due to abnormalities in iron utilization. There is increased iron transport to erythroblasts. Since the body sense anemia intestinal iron absorption increases. There is increased iron content in mitochondria of erythroblasts and systemic iron accumulation. Systemic iron overload occurs only in some forms of sideroblastic anemia, usually when the defects in iron metabolisms involve earlier stages of erythroid pathways. The development of heriditory sideroblastic anemia is the result of multiple genetic mutations in several genes involved in heme synthesis resulting in autosomal recessive congenital sideroblastic anemia. Out of many genes SLC25A38 mutations is the most common.
Pathophysiology
Physiology
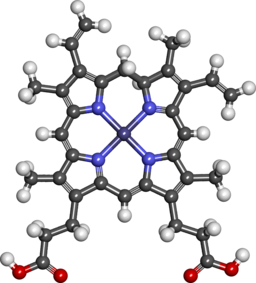 |
Heme is porphyrin containing compound, with an Fe iron ion in the centre,surrounded by heterocyclic organic ring of porphyrin.
The normal physiology of heme synthesis can be understood as follows:[1]
- Mitochondria in the developing erythroid cells are the cellular site of heme production and iron utilization.
- Glycine combines with succinylcoA to form aminolenolinic acid(ALA)
- This reaction is catalyzed by ALA sunthetase enzyme (ALAS2) in mitochondria
- ALA synthetase requires vit B-6 as a cofactor
- Two molecules of aminolenolinic acid condense in cytosol to form porphobilinogen(PBG)
- This reaction is catalyzed by zinc containing enzyme called ALA dehydratase.
- Multiple enzymatic transformations in cytoplasm produce coproporphyrinogen III (CPG).
- Coproporphyrinogen III (CPG) enters the mitochondrion.
- Additional modifications of CPG in mitochondrion produces protoporphyrin IX.
- The final step is the insertion of iron into the protoporphyrin IX ring producing heme.
- This final reaction is catalyzed by enzyme ferrochelatase.
Pathogenesis
 |
- It is understood that sideroblastic anemia is the result of defects in the steps of heme biosynthesis that occur within the mitochondrion.[2]
- Sideroblasts are the pathognomic feature of sideroblastic anemia.
- There is deffect in incorporation of iron in to heme.
- As a result the iron accumulates in mitochondria of red cell precursors
- Ring sideroblasts are erythroblasts that have iron-loaded mitochondria.
- The iron granules are arranged around the nucleus in a ring form
- They can be seen with prussian blue staining as blue granules around the nucleus.[3]
- The pathophysiology of sideroblastic anemia depends on the underlying cause.[4]
- Isoniazide, produces sideroblastic anemia in people who dont use pyridoxine prophylaxis,
- Pyridoxine is a cofactor for ALAS2 enzyme.
- Its deficiency directly impairs ALAS2 function
- Mitochondrial cytopathies
- Result from deletions of portions of the mitochondrial genome .
- The consequent marked mitochondrial dysfunction causes sideroblastic anemia in these disorders.
- Ethanol is the most common cause of toxin-induced sideroblastic anemia.
- Ethanol causes sideroblastic anemia by two mechanisms
- Direct antagonism to pyridoxal phosphate
- Dietary deficiency of this compound.
- The bone marrow shows sideroblasts and vacuoles in the normoblasts in ethanol toxicity.
- Lead intoxication causes sideroblastic anemia by inhibiting two enzymes in heme pathway:
- ALA dehaydratase
- Ferrochelatase
- Penicillamine or triethylene tetramine dihydrochloride (Trientene or TTH) used in treatment of Wilsons disease can produce sideroblastic anemia.
- Excessive chelation produces copper deficiency.
- Copper catalyzes the last step in heme biosynthesis, insertion of iron into protoporphyrin IX.
- Zinc intoxication causes sideroblastic anemia in patients using large amount of zinc supplements.
- Excessive zinc reduces serum copper levels.
Mechanism of iron overload
- Abnormalities in iron utilization in sideroblastic anemia
- There is increased iron transport to erythroblasts since the body senses anemia.
- Intestinal iron absorption increases.
- There is increased iron content in mitochondria of erythroblasts and systemic iron accumulation.
- Systemic iron overload occurs only in some forms of sideroblastic anemia, usually when the defects in iron metabolisms involve earlier stages of erythroid pathways.
Genetics
The development of sideroblastic anemia is the result of multiple genetic mutations in several genes involved in heme synthesis resulting in autosomal recessive congenital sideroblastic anemia (ARCSA)[2][4]
- SLC25A38 –SLC25A38 mutations is the most common. SLC25A38 encodes an erythroid-specific mitochondrial amino acid carrier that transports glycine into mitochondria for the first step in heme synthesis.
- GLRX5 – GLRX5 encodes protein used in the synthesis of iron-sulfur (Fe-S) clusters.
- HSPA9 – HSPA9 encodes the mitochondrial HSP70 homologue HSPA9, which is also involved in Fe-S cluster formation.
- FECH – FECH encodes ferrochelatase, the final enzyme in the heme synthesis pathway, which inserts an iron atom into protoporphyrin IX
- X-linked sideroblastic anemia with ataxia (ABCB7 mutation)
- Pearson syndrome ( mitochondrial DNA deletion)
- Pearson syndrome is a congenital condition
- It affects multiple systems,
- Results in severe sideroblastic anemia, thrombocytopenia and neutropenia.
- Pancreatic insufficiency, lactic acidosis, and growth retardation are other common features.
References
- ↑ Layer G, Reichelt J, Jahn D, Heinz DW (June 2010). "Structure and function of enzymes in heme biosynthesis". Protein Sci. 19 (6): 1137–61. doi:10.1002/pro.405. PMC 2895239. PMID 20506125.
- ↑ 2.0 2.1 Fleming MD (2011). "Congenital sideroblastic anemias: iron and heme lost in mitochondrial translation". Hematology Am Soc Hematol Educ Program. 2011: 525–31. doi:10.1182/asheducation-2011.1.525. PMID 22160084.
- ↑ Cazzola M, Invernizzi R (June 2011). "Ring sideroblasts and sideroblastic anemias". Haematologica. 96 (6): 789–92. doi:10.3324/haematol.2011.044628. PMC 3105636. PMID 21632840.
- ↑ 4.0 4.1 Fujiwara T, Harigae H (December 2013). "Pathophysiology and genetic mutations in congenital sideroblastic anemia". Pediatr Int. 55 (6): 675–9. doi:10.1111/ped.12217. PMID 24003969.