Severe acute respiratory syndrome pathophysiology
|
Severe Acute Respiratory Syndrome Microchapters |
|
Differentiating Severe Acute Respiratory Syndrome from other Diseases |
|---|
|
Diagnosis |
|
Treatment |
|
Case Studies |
|
Severe acute respiratory syndrome pathophysiology On the Web |
|
American Roentgen Ray Society Images of Severe acute respiratory syndrome pathophysiology |
|
Severe acute respiratory syndrome pathophysiology in the news |
|
Directions to Hospitals Treating Severe acute respiratory syndrome |
|
Risk calculators and risk factors for Severe acute respiratory syndrome pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Pathophysiology
SARS Coronavirus
Coronaviruses are positive-strand, enveloped RNA viruses that are important pathogens of mammals and birds. This group of viruses cause enteric or respiratory tract infections in a variety of animals including humans, livestock and pets.[1]
Initial electron microscopic examination in Hong Kong and Germany found viral particles with structures suggesting paramyxovirus in respiratory secretions of SARS patients. Subsequently, in Canada, electron microscopic examination found viral particles with structures suggestive of metapneumovirus (a subtype of paramyxovirus) in respiratory secretions. Chinese researchers also reported that a chlamydia-like disease may be behind SARS. The Pasteur Institute in Paris identified coronavirus in samples taken from six patients, so did the laboratory of Malik Peiris at the University of Hong Kong, which in fact was the first to announce (on March 21, 2003) the discovery of a new coronavirus as the possible cause of SARS after successfully cultivating it from tissue samples and was also amongst the first to develop a test for the presence of the virus. The CDC noted viral particles in affected tissue (finding a virus in tissue rather than secretions suggests that it is actually pathogenic rather than an incidental finding). Upon electron microscopy, these tissue viral inclusions resembled coronaviruses, and comparison of viral genetic material obtained by PCR with existing genetic libraries suggested that the virus was a previously unrecognized coronavirus. Sequencing of the virus genome — which computers at the British Columbia Cancer Agency in Vancouver completed at 4 a.m. Saturday, April 12, 2003 — was the first step toward developing a diagnostic test for the virus, and possibly a vaccine.[2] A test was developed for antibodies to the virus, and it was found that patients did indeed develop such antibodies over the course of the disease, which is highly suggestive of a causative role.
On April 16, 2003, the WHO issued a press release stating that a coronavirus identified by a number of laboratories was the official cause of SARS.[3] Scientists at Erasmus University in Rotterdam, the Netherlands demonstrated that the SARS coronavirus fulfilled Koch's postulates thereby confirming it as the causative agent. In the experiments, macaques infected with the virus developed the same symptoms as human SARS victims.[4]
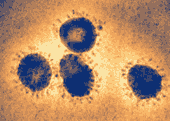
An article published in The Lancet identifies a coronavirus as the probable causative agent.
In late May 2003, studies from samples of wild animals sold as food in the local market in Guangdong, China found that the SARS coronavirus could be isolated from civets. This suggests that the SARS virus crossed the xenographic barrier from civets. In 2005, two studies identified a number of SARS-like coronaviruses in Chinese bats.[5][6] Phylogenetic analysis of these viruses indicated a high probability that SARS coronavirus originated in bats and spread to humans either directly, or through civet cats. The bats did not show any visible signs of disease.
Viral Replication
Coronavirus (CoV) genome replication takes place in the cytoplasm in a membrane-protected microenvironment and starts with the translation of the genome to produce the viral replicase. CoV transcription involves a discontinuous RNA synthesis (template switch) during the extension of a negative copy of the subgenomic mRNAs. The requirement for base pairing during transcription has been formally demonstrated in arteriviruses and CoVs. CoV N protein is required for coronavirus RNA synthesis and has RNA chaperone activity that may be involved in template switch. Both viral and cellular proteins are required for replication and transcription. CoVs initiate translation by cap-dependent and cap-independent mechanisms. Cell macromolecular synthesis may be controlled after CoV infection by locating some virus proteins in the host cell nucleus. Infection by different coronaviruses cause in the host alteration in the transcription and translation patterns, in the cell cycle, the cytoskeleton, apoptosis and coagulation pathways, inflammation and immune and stress responses. The balance between genes up- and down-regulated could explain the pathogenesis caused by these viruses. Coronavirus expression systems based on single genome constructed by targeted recombination, or by using infectious cDNAs, have been developed. The possibility of expressing different genes under the control of transcription regulating sequences (TRSs) with programmable strength and engineering tissue and species tropism indicates that CoV vectors are flexible. CoV based vectors have emerged with high potential vaccine development and possibly for gene therapy.[7]
References
- ↑ Thiel V (editor). (2007). Coronaviruses: Molecular and Cellular Biology (1st ed. ed.). Caister Academic Press. ISBN 978-1-904455-16-5 .
- ↑ Marra, MA, Jones, SJ, Astell, CR et al (2003) The genome sequence of the SARS-associated coronavirus. Science 300(5624):1377–1378
- ↑ Coronavirus never before seen in humans is the cause of SARS, New York: United Nations World Health Organization, 16 April 2006. URL Accessed 5 July 2006.
- ↑ Fouchier, RAM, Kuiken, T, Schutten, M et al (2003). Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 423:240.
- ↑ Li, W, Shi, A, Yu, M et al (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676–679.
- ↑ Lau, SKP, Woo, PCY, Li, KSM et al. (2005). Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences 102(29):14040–14045.
- ↑ Enjuanes L; et al. (2008). "Coronavirus Replication and Interaction with Host". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.