Lipopolysaccharide
Lipopolysaccharide (LPS) is a large molecule consisting of a lipid and a polysaccharide (carbohydrate) joined by a covalent bond.
Functions
LPS is a major component of the outer membrane of Gram-negative bacteria, contributing greatly to the structural integrity of the bacteria, and protecting the membrane from certain kinds of chemical attack. LPS is an endotoxin, and induces a strong response from normal animal immune systems. The only Gram-positive bacteria that possesses LPS is Listeria monocytogenes, the common infective agent in unpasteurized milk.
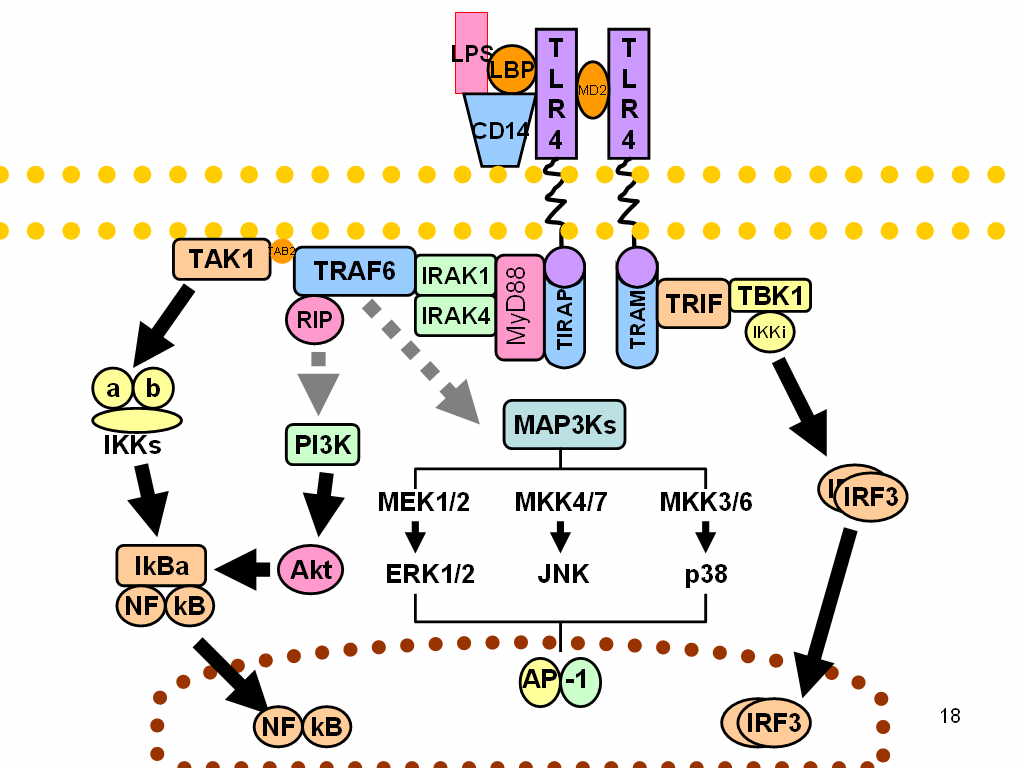
LPS acts as the prototypical endotoxin, because it binds the CD14/TLR4/MD2 receptor complex, which promotes the secretion of pro-inflammatory cytokines in many cell types, but especially in macrophages. An "LPS challenge" in immunology is the exposing of the subject to an LPS which may act as a toxin.
LPS also increases the negative charge of the cell membrane and helps stabilize the overall membrane structure.
Composition
It comprises three parts:
- polysaccharide (O) side chains
- core polysaccharide (core oligosaccharide in genus Neisseria)
- lipid A
Lipid A
Lipid A contains unusual fatty acids (e.g. hydroxy-myristic acid) and is embedded into the outer membrane while the rest of the LPS projects from the surface. This is the key in the toxicity.
Core oligosaccharide
Core oligosaccharide contains unusual sugars (e.g. KDO, keto-deoxyoctulonate and heptose).
The core oligosaccharide is attached to lipid A, which is also in part responsible for the toxicity of gram-negative bacteria.
O-antigen
The polysaccharide side chain is referred to as the O-antigen of the bacteria. O side chain (O-antigen) is also a polysaccharide chain that extends from the core polysaccharide. The composition of the O side chain varies between different gram-negative bacterial strains. The presence or absence of O chains determine whether the LPS is considered rough or smooth. Full length O-chains would render the LPS smooth while the absence or reduction of O-chains would make the LPS rough. [1]Bacteria with rough LPS usually have more penetrable cell membranes to hydrophobic antibiotics since a rough LPS is more hydrophobic. [2]
O side chains are easily recognized by the antibodies of the host, however, the nature of the chain can easily be modified by Gram-negative bacteria to avoid detection. The structure of the core and the O-antigen is often determined by methylation analysis or NMR-spectroscopy.
LPS modifications
The making of LPS can be modified in order to present a specific sugar structure. Those can be recognised by either other LPS (which enables to inhibit LPS toxins) or glycosyltransferases which use those sugar structure to add more specific sugars.
Variability and effect upon specificity
O-antigens (the outer carbohydrates) are the most variable portion of the LPS molecule, imparting the antigenic specificity. In contrast, lipid A is the most conserved part. However, —lipid A composition also may vary (e.g., in number and nature of acyl chains even within or between genera). Some of these variations may impart antagonistic properties to these LPS. For example Rhodobacter sphaeroides diphosphoryl lipid A (RsDPLA) is a potent antagonist of LPS in human cells, but is an agonist in hamster and equine cells.
It has been speculated that conical Lipid A (eg from E coli) are more agonistic, less conical lipid A like those of Porphyromonas gingivalis may activate a different signal (TLR2 instead of TLR4), and completely cylindrical lipid A like that of Rhodobacter sphaeroides is antagonistic to TLRs.[3][4]
Functions
LPS function has been under experimental research for several years due to its role in activating many transcription factors. LPS also produces many types of mediators involved in septic shock.
Diversity
Lipololysaccharide gene clusters are highly variable between different strains, subspecies, species of bacterial pathogens of plants and animals.[5][6]
References
- ↑ Rittig MG; et al. (2004). "Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes". Journal of Leukocyte Biology. 5 (4): 196–200. PMID 12960272.
- ↑ Tsujimoto H; et al. (2003). "Diffusion of macrolide antibiotics through the outer membrane of Moraxella catarrhalis". Journal of Infection and Chemotherapy. 74: 1045–1055. PMID 11810516.
- ↑ Netea M; et al. (2002). "Does the shape of lipid A determine the interaction of LPS with Toll-like receptors?". Trends Immunol. 23 (3): 135–9. PMID 11864841.
- ↑ Seydel U, Oikawa M, Fukase K, Kusumoto S, Brandenburg K (2000). "Intrinsic conformation of lipid A is responsible for agonistic and antagonistic activity". Eur J Biochem. 267 (10): 3032–9. PMID 10806403.
- ↑ Reeves P, Wang L (2002). "Genomic organization of LPS-specific loci". Curr Top Microbiol Immunol. 264 (1): 109–35. PMID 12014174.
- ↑ Patil P, Sonti R (2004). "Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice". BMC Microbiol. 4 (1): 40. PMID 15473911.
See also
External links
- Lipopolysaccharides at the US National Library of Medicine Medical Subject Headings (MeSH)
de:Lipopolysaccharide it:Lipopolisaccaride he:ליפופוליסכריד sv:Lipopolysackarider uk:Ліпополісахариди Template:WH Template:WS