HLA E
| Major histocompatibility complex, class I, E | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 PDB rendering based on 1kpr. | |||||||||||||
| |||||||||||||
| Identifiers | |||||||||||||
| Symbols | HLA-E ; DKFZp686P19218; EA1.2; EA2.1; HLA-6.2; MHC | ||||||||||||
| External IDs | Template:OMIM5 Template:MGI HomoloGene: 38064 | ||||||||||||
| |||||||||||||
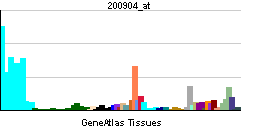
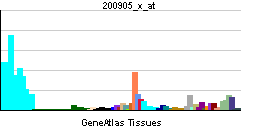
| RNA expression pattern | |||||||||||||
 | |||||||||||||
 | |||||||||||||
 | |||||||||||||
| More reference expression data | |||||||||||||
| Orthologs | |||||||||||||
| Template:GNF Ortholog box | |||||||||||||
| Species | Human | Mouse | |||||||||||
| Entrez | n/a | n/a | |||||||||||
| Ensembl | n/a | n/a | |||||||||||
| UniProt | n/a | n/a | |||||||||||
| RefSeq (mRNA) | n/a | n/a | |||||||||||
| RefSeq (protein) | n/a | n/a | |||||||||||
| Location (UCSC) | n/a | n/a | |||||||||||
| PubMed search | n/a | n/a | |||||||||||
Major histocompatibility complex, class I, E, also known as HLA-E, is a human gene.
HLA-E belongs to the HLA class I heavy chain paralogues. This class I molecule is a heterodimer consisting of a heavy chain and a light chain (beta-2 microglobulin). The heavy chain is anchored in the membrane. HLA-E binds a restricted subset of peptides derived from the leader peptides of other class I molecules. The heavy chain is approximately 45 kDa and its gene contains 8 exons. Exon one encodes the leader peptide, exons 2 and 3 encode the alpha1 and alpha2 domains, which both bind the peptide, exon 4 encodes the alpha3 domain, exon 5 encodes the transmembrane region, and exons 6 and 7 encode the cytoplasmic tail.[1]
HLA-E is a protein that is a member of the HLA family. It is one of a family of molecules known as MHC class Ib and has a very specialized role in cell recognition by NK cells (Natural Killer). HLA-E is very highly conserved and presents a small repertoire of peptides of various origins.[2] HLA-E is expressed ubiquitously. The primary peptides HLA-E binds are derived from the leader sequence of other HLA class Ia molecules, such as HLA-A, B, C, G.These proteins bind to the Peptide binding groove of the HLA-E molecule, and this allows it to be expressed on on the surface. NK cells recognise bound HLA-E+Peptide combination using the heterodimeric inhibitory receptor CD94/NKG2A or the activating receptor NKG2C/CD94 . When CD94/NKG2A is stimulated, it produces an inhibitory effect on the cytotoxic activity of the NK cell to prevent cell lysis.
A number of viruses have evolved to express proteins that inhibit the expression of MHC I molecules on cell surface of cells they infect, thereby preventing Antigen presentation that would allow the Immune system to recognise infection. NK cells therefore identify cells that are exhibiting such characteristics of viral infection.
HLA E is also reported to interact with the T-cell receptor.[3] HLA-E presents a peptide (VMAPRTLIL) derived from the UL40 protein from cytomegalovirus. This peptide is highly similar to the leader peptides derived from other HLA molecules and acts as a surrogate "leader peptide" and interacts with CD94/NKG2 receptors. This engagement relays into an overall inhibitory effect on NK-cell meditated cytotoxicity.
References
- ↑ "Entrez Gene: HLA-E major histocompatibility complex, class I, E".
- ↑ Rodgers JR, Cook, RG (2005). "MHC CLASS IB MOLECULES BRIDGE INNATE AND ACQUIRED IMMUNITY". Nat Rev Immunol. 5 (6): 459–471. PMID 15928678.
- ↑ Hoare HL, Sullivan LC, Pietra G; et al. (2006). "Structural basis for a major histocompatibility complex class Ib-restricted T cell response". Nat. Immunol. 7 (3): 256–64. doi:10.1038/ni1312. PMID 16474394.
Further reading
- Arnaiz-Villena A, Martinez-Laso J, Alvarez M; et al. (1997). "Primate Mhc-E and -G alleles". Immunogenetics. 46 (4): 251–66. PMID 9218527.
- Geyer M, Fackler OT, Peterlin BM (2001). "Structure--function relationships in HIV-1 Nef". EMBO Rep. 2 (7): 580–5. doi:10.1093/embo-reports/kve141. PMID 11463741.
- Greenway AL, Holloway G, McPhee DA; et al. (2004). "HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication". J. Biosci. 28 (3): 323–35. PMID 12734410.
- Bénichou S, Benmerah A (2003). "[The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway]". Med Sci (Paris). 19 (1): 100–6. PMID 12836198.
- Leavitt SA, SchOn A, Klein JC; et al. (2004). "Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm". Curr. Protein Pept. Sci. 5 (1): 1–8. PMID 14965316.
- Tolstrup M, Ostergaard L, Laursen AL; et al. (2004). "HIV/SIV escape from immune surveillance: focus on Nef". Curr. HIV Res. 2 (2): 141–51. PMID 15078178.
- Joseph AM, Kumar M, Mitra D (2005). "Nef: "necessary and enforcing factor" in HIV infection". Curr. HIV Res. 3 (1): 87–94. PMID 15638726.
- Anderson JL, Hope TJ (2005). "HIV accessory proteins and surviving the host cell". Current HIV/AIDS reports. 1 (1): 47–53. PMID 16091223.
- Kozlowski S, Corr M, Takeshita T; et al. (1992). "Serum angiotensin-1 converting enzyme activity processes a human immunodeficiency virus 1 gp160 peptide for presentation by major histocompatibility complex class I molecules". J. Exp. Med. 175 (6): 1417–22. PMID 1316930.
- Ulbrecht M, Honka T, Person S; et al. (1992). "The HLA-E gene encodes two differentially regulated transcripts and a cell surface protein". J. Immunol. 149 (9): 2945–53. PMID 1401923.
- Truong MJ, Gruart V, Capron A; et al. (1992). "Cloning and expression of a cDNA encoding a non-classical MHC class I antigen (HLA-E) in eosinophils from hypereosinophilic patients". J. Immunol. 148 (2): 627–32. PMID 1530866.
- Houlihan JM, Biro PA, Fergar-Payne A; et al. (1992). "Evidence for the expression of non-HLA-A,-B,-C class I genes in the human fetal liver". J. Immunol. 149 (2): 668–75. PMID 1624808.
- Ohya K, Kondo K, Mizuno S (1990). "Polymorphism in the human class I MHC locus HLA-E in Japanese". Immunogenetics. 32 (3): 205–9. PMID 1977695.
- Wei XH, Orr HT (1991). "Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue". Hum. Immunol. 29 (2): 131–42. PMID 2249951.
- Srivastava R, Chorney MJ, Lawrance SK; et al. (1987). "Structure, expression, and molecular mapping of a divergent member of the class I HLA gene family". Proc. Natl. Acad. Sci. U.S.A. 84 (12): 4224–8. PMID 2438694.
- Pohla H, Kuon W, Tabaczewski P; et al. (1989). "Allelic variation in HLA-B and HLA-C sequences and the evolution of the HLA-B alleles". Immunogenetics. 29 (5): 297–307. PMID 2714852.
- Takahashi H, Merli S, Putney SD; et al. (1989). "A single amino acid interchange yields reciprocal CTL specificities for HIV-1 gp160". Science. 246 (4926): 118–21. PMID 2789433.
- Carroll MC, Katzman P, Alicot EM; et al. (1988). "Linkage map of the human major histocompatibility complex including the tumor necrosis factor genes". Proc. Natl. Acad. Sci. U.S.A. 84 (23): 8535–9. PMID 2825194.
- Mizuno S, Trapani JA, Koller BH; et al. (1988). "Isolation and nucleotide sequence of a cDNA clone encoding a novel HLA class I gene". J. Immunol. 140 (11): 4024–30. PMID 3131426.
- Koller BH, Geraghty DE, Shimizu Y; et al. (1988). "HLA-E. A novel HLA class I gene expressed in resting T lymphocytes". J. Immunol. 141 (3): 897–904. PMID 3260916.
| Stub icon | This biochemistry article is a stub. You can help Wikipedia by expanding it. |