Basal cell carcinoma pathophysiology: Difference between revisions
| (25 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{Basal cell carcinoma}} | {{Basal cell carcinoma}} | ||
{{CMG}};{{AE}}{{ | {{CMG}};{{AE}}{{M.N}} | ||
==Overview== | ==Overview== | ||
Basal cell | Basal cell carcinoma is one of the most common [[skin cancers]]. It is commonly known as [[rodent ulcer]] due to its distinct [[Morphology (biology)|morphology]] characterized by pearly pink [[nodules]] with [[telangiectasias]], rolled borders, and central crusting with or without an [[Ulceration|ulcerating]] [[lesion]]. The majority common [[Causes|cause]] for the [[development]] of the basal cell carcinoma involves [[radiation exposure]] and [[mutations]] that involve many [[genes]] including sonic [[Hedgehog (cell signaling)|hedgehog]] [[gene]], [[PTCH1]] [[gene]], and other [[Gain-of-function mutation|gain-of-function mutations]] which further depend on the subtypes such as [[nodular]], [[superficial]], Infundibulocystic, [[fibroepithelial]], morpheaform, infiltrative, micronodular, and basosquamous basal cell carcinomas. | ||
==Pathophysiology== | ==Pathophysiology== | ||
===Pathogenesis=== | |||
The exact pathogenesis of basal cell carcinoma is not completely understood | |||
===Genetics=== | ===Genetics=== | ||
*A [[number]] of aberrations involving the [[sonic hedgehog]] [[signaling pathway]](SHH) are noted<ref name="pmid24587976">{{cite journal |vauthors=Mohan SV, Chang AL |title=Advanced Basal Cell Carcinoma: Epidemiology and Therapeutic Innovations |journal=Curr Dermatol Rep |volume=3 |issue= |pages=40–45 |date=2014 |pmid=24587976 |pmc=3931971 |doi=10.1007/s13671-014-0069-y |url=}}</ref><ref name="pmid29165358">{{cite journal |vauthors=Pellegrini C, Maturo MG, Di Nardo L, Ciciarelli V, Gutiérrez García-Rodrigo C, Fargnoli MC |title=Understanding the Molecular Genetics of Basal Cell Carcinoma |journal=Int J Mol Sci |volume=18 |issue=11 |pages= |date=November 2017 |pmid=29165358 |pmc=5713451 |doi=10.3390/ijms18112485 |url=}}</ref><ref name="pmid30405815">{{cite journal |vauthors=Yunoki T, Tabuchi Y, Hirano T, Miwa S, Imura J, Hayashi A |title=Gene networks in basal cell carcinoma of the eyelid, analyzed using gene expression profiling |journal=Oncol Lett |volume=16 |issue=5 |pages=6729–6734 |date=November 2018 |pmid=30405815 |pmc=6202553 |doi=10.3892/ol.2018.9484 |url=}}</ref> | The development of basal cell carcinoma is the result of multiple genetic mutations such as sonic hedgehog pathway mutations, and PTCH1 gene mutations | ||
*This pathway is [[vital]] for the [[Regulation of gene expression|regulation]] of [[cell growth]], and [[differentiation]] and loss of [[inhibition]] of this pathway is associated with [[development]] of basal cell cancer | *A [[number]] of aberrations involving the [[sonic hedgehog]] [[signaling pathway]](SHH) are noted.<ref name="pmid24587976">{{cite journal |vauthors=Mohan SV, Chang AL |title=Advanced Basal Cell Carcinoma: Epidemiology and Therapeutic Innovations |journal=Curr Dermatol Rep |volume=3 |issue= |pages=40–45 |date=2014 |pmid=24587976 |pmc=3931971 |doi=10.1007/s13671-014-0069-y |url=}}</ref><ref name="pmid29165358">{{cite journal |vauthors=Pellegrini C, Maturo MG, Di Nardo L, Ciciarelli V, Gutiérrez García-Rodrigo C, Fargnoli MC |title=Understanding the Molecular Genetics of Basal Cell Carcinoma |journal=Int J Mol Sci |volume=18 |issue=11 |pages= |date=November 2017 |pmid=29165358 |pmc=5713451 |doi=10.3390/ijms18112485 |url=}}</ref><ref name="pmid30405815">{{cite journal |vauthors=Yunoki T, Tabuchi Y, Hirano T, Miwa S, Imura J, Hayashi A |title=Gene networks in basal cell carcinoma of the eyelid, analyzed using gene expression profiling |journal=Oncol Lett |volume=16 |issue=5 |pages=6729–6734 |date=November 2018 |pmid=30405815 |pmc=6202553 |doi=10.3892/ol.2018.9484 |url=}}</ref><ref name="pmid26029015">{{cite journal |vauthors=Marzuka AG, Book SE |title=Basal cell carcinoma: pathogenesis, epidemiology, clinical features, diagnosis, histopathology, and management |journal=Yale J Biol Med |volume=88 |issue=2 |pages=167–79 |date=June 2015 |pmid=26029015 |pmc=4445438 |doi= |url=}}</ref> | ||
*The majority of [[mutations]] in sporadic basal cell carcinoma and [[basal cell nevus syndrome]](BCNS) [[patients]] occur in [[PTCH1]] [[gene]], a [[protein]] that inhibits smoothened [[gene]](SMO) | *This pathway is [[vital]] for the [[Regulation of gene expression|regulation]] of [[cell growth]], and [[differentiation]] and loss of [[inhibition]] of this pathway is associated with [[development]] of basal cell cancer. | ||
*The [[second]] most common [[mutation]] in sporadic basal cell carcinoma and [[basal cell nevus syndrome]](BCNS) [[patients]] are [[Gain-of-function mutation|gain-of-function]] [[mutations]] of the smoothened [[gene]](SMO) | *The majority of [[mutations]] in sporadic basal cell carcinoma and [[basal cell nevus syndrome]](BCNS) [[patients]] occur in [[PTCH1]] [[gene]], a [[protein]] that inhibits smoothened [[gene]] (SMO). | ||
*Loss of [[PTCH1]] results in the failure of Smoothened [[inhibition]], subsequently leading to increases in [[GLI1]] levels, changes in [[transcription]], and subsequent [[tumorigenesis]] | *The [[second]] most common [[mutation]] in sporadic basal cell carcinoma and [[basal cell nevus syndrome]](BCNS) [[patients]] are [[Gain-of-function mutation|gain-of-function]] [[mutations]] of the smoothened [[gene]] (SMO). | ||
*Loss of [[PTCH1]] results in the failure of Smoothened [[inhibition]], subsequently leading to increases in [[GLI1]] levels, changes in [[transcription]], and subsequent [[tumorigenesis]]. | |||
*[[Gain-of-function mutation|Gain-of-function]] smoothened(SMO) [[mutations]] also leads to increased [[GLI1]] levels and [[tumorigenesis]] | *[[Gain-of-function mutation|Gain-of-function]] smoothened(SMO) [[mutations]] also leads to increased [[GLI1]] levels and [[tumorigenesis]] | ||
{{Family tree/start}} | {{Family tree/start}} | ||
| Line 29: | Line 31: | ||
{{Family tree | | | | E01 | | | |E01= Tumorigenesis}} | {{Family tree | | | | E01 | | | |E01= Tumorigenesis}} | ||
{{Family tree/end}} | {{Family tree/end}} | ||
[[File:Soinic hedgehog pathway signalling.jpg|thumb|500px|none|Sonic hedgehog signaling pathway. SHH ligand binds to and inhibits the PTCH transmembrane protein. The inhibition of PTCH relieves suppression of / (Smoothened), which then activates the GLI transcription factors. The GLI proteins translocate from the cytoplasm to the nucleus, where they drive gene transcription. (Courtesy of Alexander G. Marzuka, MD),https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4445438/]] | |||
Other Genetic Changes: | |||
*Point mutations in the TP53 gene, the tumor supressor gene are the second most common genetic alteration noticed in BCCs | |||
*Some mutations in the CDKN2A locus and in ras gene family (H-ras, K-ras, and N-ras) are also identified in a smaller proportion of sporadic BCCs | |||
===Enviromental Exposure=== | ===Enviromental Exposure=== | ||
*Basal cell carcinomas develop in the [[basal cell layer]] of the [[skin]] | *Basal cell carcinomas develop in the [[basal cell layer]] of the [[skin]].<ref name="pmid28954101">{{cite journal |vauthors=Montagna E, Lopes OS |title=Molecular basis of basal cell carcinoma |journal=An Bras Dermatol |volume=92 |issue=4 |pages=517–520 |date=2017 |pmid=28954101 |pmc=5595599 |doi=10.1590/abd1806-4841.20176544 |url=}}</ref> | ||
*Cumulative [[DNA damage]] caused by [[chronic]] [[sunlight]] exposure results in [[DNA mutations]] that predispose to the [[development]] of basal cell carcinoma | *Cumulative [[DNA damage]] caused by [[chronic]] [[sunlight]] exposure results in [[DNA mutations]] that predispose to the [[development]] of basal cell carcinoma. | ||
*While [[DNA repair]] eliminates most [[Ultraviolet|UV-]]<nowiki/>induced damage, not all cross-links are excised, which eventually results in [[mutation]]s | *While [[DNA repair]] eliminates most [[Ultraviolet|UV-]]<nowiki/>induced damage, not all cross-links are excised, which eventually results in [[mutation]]s. | ||
*Apart from the [[mutagenesis]], [[sunlight]] [[depresses]] the local [[immune system]], possibly decreasing [[immune]] surveillance for [[new]] [[Tumor cell|tumor cells]] | *Apart from the [[mutagenesis]], [[sunlight]] [[depresses]] the local [[immune system]], possibly decreasing [[immune]] surveillance for [[new]] [[Tumor cell|tumor cells]]. | ||
=== | ===Gross and microscopic pathology=== | ||
Basal cell carcinoma [[pathological]] features mainly depend upon the subtype. The following table summarizes them:<ref name="CameronLee2019">{{cite journal|last1=Cameron|first1=Michael C.|last2=Lee|first2=Erica|last3=Hibler|first3=Brian P.|last4=Barker|first4=Christopher A.|last5=Mori|first5=Shoko|last6=Cordova|first6=Miguel|last7=Nehal|first7=Kishwer S.|last8=Rossi|first8=Anthony M.|title=Basal cell carcinoma|journal=Journal of the American Academy of Dermatology|volume=80|issue=2|year=2019|pages=303–317|issn=01909622|doi=10.1016/j.jaad.2018.03.060}}</ref><ref name="pmid25134314">{{cite journal |vauthors=Sehgal VN, Chatterjee K, Pandhi D, Khurana A |title=Basal cell carcinoma: pathophysiology |journal=Skinmed |volume=12 |issue=3 |pages=176–81 |date=2014 |pmid=25134314 |doi= |url=}}</ref> | *On gross and microscopic histopathological analysis the characteristic findings of basal cell carcinoma are described as below: | ||
*Basal cell carcinoma [[pathological]] features mainly depend upon the subtype. The following table summarizes them:<ref name="CameronLee2019">{{cite journal|last1=Cameron|first1=Michael C.|last2=Lee|first2=Erica|last3=Hibler|first3=Brian P.|last4=Barker|first4=Christopher A.|last5=Mori|first5=Shoko|last6=Cordova|first6=Miguel|last7=Nehal|first7=Kishwer S.|last8=Rossi|first8=Anthony M.|title=Basal cell carcinoma|journal=Journal of the American Academy of Dermatology|volume=80|issue=2|year=2019|pages=303–317|issn=01909622|doi=10.1016/j.jaad.2018.03.060}}</ref><ref name="pmid25134314">{{cite journal |vauthors=Sehgal VN, Chatterjee K, Pandhi D, Khurana A |title=Basal cell carcinoma: pathophysiology |journal=Skinmed |volume=12 |issue=3 |pages=176–81 |date=2014 |pmid=25134314 |doi= |url=}}</ref> | |||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| rowspan="2" align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Subtypes of BCC'''}} | |||
| colspan="2" align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Gross features'''}} | |||
| colspan="2" align="center" style="background: #4479BA;" | {{fontcolor|#FFF|''' Microscopic features'''}} | |||
|- | |- | ||
| align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Findings'''}} | |||
| align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Images'''}} | |||
| align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Findings'''}} | |||
| align="center" style="background: #4479BA;" | {{fontcolor|#FFF|'''Images'''}} | |||
|- | |- | ||
| [[Nodular]] | | [[Nodular]] | ||
| Line 58: | Line 65: | ||
[[File:BCC Nodular type.jpg|thumb|center|150px|M. Sand, D. Sand, C. Thrandorf, V. Paech, P. Altmeyer, F. G. Bechara [CC BY 2.0 (https://creativecommons.org/licenses/by/2.0)], via Wikimedia Commons]] | [[File:BCC Nodular type.jpg|thumb|center|150px|M. Sand, D. Sand, C. Thrandorf, V. Paech, P. Altmeyer, F. G. Bechara [CC BY 2.0 (https://creativecommons.org/licenses/by/2.0)], via Wikimedia Commons]] | ||
| | | | ||
*Discrete nests of [[malignant]] basaloid [[cells]] in the [[dermis]] | *[[Discrete distribution|Discrete]] nests of [[malignant]] basaloid [[cells]] in the [[dermis]] | ||
*Peripheral palisading | *Peripheral palisading | ||
*Mucoid [[stroma]] containing plump [[spindle cells]] | *Mucoid [[stroma]] containing plump [[spindle cells]] | ||
| Line 82: | Line 89: | ||
| | | | ||
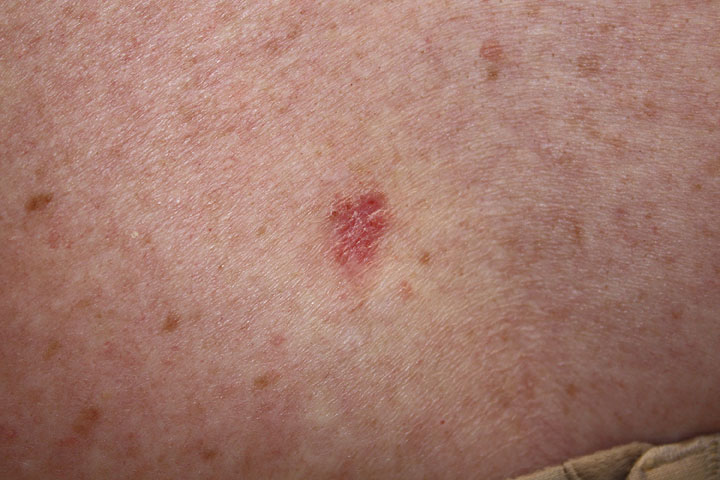
*Well-circumscribed pearly papule | *Well-circumscribed pearly [[papule]] | ||
*Most common on the head and neck region | *Most common on the [[head]] and [[neck]] region | ||
| | | | ||
| | | | ||
*Well-circumscribed | *Well-circumscribed | ||
*Anastomosing strands of basaloid cells and scattered infundibulum-like cystic structures | *[[Anastomosing]] strands of basaloid [[Cells (biology)|cells]] and scattered [[infundibulum]]-like [[cystic]] structures | ||
| | | | ||
|- | |- | ||
| Line 94: | Line 101: | ||
| | | | ||
*Skin-colored/erythematous | *[[Skin]]-colored/[[erythematous]] | ||
*Sessile plaque/pedunculated papulonodule | *[[Sessile]] [[plaque]]/[[pedunculated]] papulonodule | ||
*They have a predilection for trunk region | *They have a predilection for [[trunk]] region | ||
| | | | ||
| | | | ||
*Multiple collections of delicate strands of epidermal basaloid keratinocytes | *Multiple collections of delicate strands of [[epidermal]] basaloid [[keratinocytes]] | ||
*These are usually arranged in a reticular pattern within a spindle cell stroma | *These are usually arranged in a [[reticular]] pattern within a [[spindle cell]] [[stroma]] | ||
| | | | ||
|- | |- | ||
| Line 107: | Line 114: | ||
| | | | ||
*Infiltrated plaque with poorly defined borders and shiny surface | *Infiltrated [[plaque]] with poorly defined borders and shiny [[Surface area|surface]] | ||
*Most common on head and neck region | *Most common on [[head]] and [[neck]] region | ||
| | | | ||
[[File:PMC3339125 JCAS-5-3-g004.png|thumb|center|150px|Dermatology Centre, Salford Royal Hospital, NHS Foundation Trust, Stott Lane, Salford M6 8HD, UK.]] | [[File:PMC3339125 JCAS-5-3-g004.png|thumb|center|150px|Dermatology Centre, Salford Royal Hospital, NHS Foundation Trust, Stott Lane, Salford M6 8HD, UK.]] | ||
| | | | ||
*Thin cords of basaloid cells surrounded by a sclerotic collagenous stroma | *Thin cords of basaloid [[Cells (biology)|cells]] surrounded by a sclerotic [[collagenous]] [[stroma]] | ||
*Absent peripheral palisading and stromal cleft formation | *Absent peripheral palisading and [[stromal]] [[cleft]] formation | ||
*Positive staining of tumor stroma with smooth muscle alpha-actin | *Positive staining of [[tumor]] [[stroma]] with [[smooth muscle]] [[alpha-actin]] | ||
| | | | ||
[[File:PMC4513413 IDOJ-6-286-g002.png|thumb|center|200px|Department of Pathology, Columbia University Medical Center, New York, USA]] | [[File:PMC4513413 IDOJ-6-286-g002.png|thumb|center|200px|Department of Pathology, Columbia University Medical Center, New York, USA]] | ||
| Line 123: | Line 130: | ||
| | | | ||
*Poorly defined | *Poorly defined | ||
*Indurated, flat or depressed plaque with white, yellow, or pale pink color | *[[Induration|Indurated]], flat or depressed [[plaque]] with white, yellow, or pale pink color | ||
*They may have overlying crusts, erosions, ulcerations, or papules | *They may have overlying crusts, erosions, [[ulcerations]], or [[papules]] | ||
| | | | ||
[[File:Basal cell carcinoma (1).jpg|thumb|center|200px|Kelly Nelson (Photographer) [Public domain], via Wikimedia Commons,https://upload.wikimedia.org/wikipedia/commons/9/9b/Basal_cell_carcinoma_%281%29.jpg,]] | [[File:Basal cell carcinoma (1).jpg|thumb|center|200px|Kelly Nelson (Photographer) [Public domain], via Wikimedia Commons,https://upload.wikimedia.org/wikipedia/commons/9/9b/Basal_cell_carcinoma_%281%29.jpg,]] | ||
| | | | ||
*Thin cords with angulated ends of few basaloid keratinocytes | *Thin cords with angulated ends of few basaloid [[keratinocytes]] | ||
*Usually embedded in a classic mucinous/myxoid stroma | *Usually embedded in a classic [[mucinous]]/myxoid [[stroma]] | ||
| | | | ||
|- | |- | ||
| Line 136: | Line 143: | ||
| | | | ||
*Erythematous macule or thin papule/plaque | *[[Erythematous]] [[macule]] or thin [[papule]]/[[plaque]] | ||
| | | | ||
| | | | ||
*Multiple small aggregates of basaloid cells within the dermis, with subtle peripheral palisading and retraction artifact | *Multiple small aggregates of basaloid [[Cells (biology)|cells]] within the [[dermis]], with subtle peripheral palisading and retraction artifact | ||
| | | | ||
|- | |- | ||
| Line 146: | Line 153: | ||
| | | | ||
*Majority found on the head and neck | *Majority found on the [[head]] and [[neck]] | ||
| | | | ||
| | | | ||
*Well-defined nodular or superficial BCC component overlying an invasive front showing basal cell carcinoma and squamous cell carcinoma histologic feature | *Well-defined [[nodular]] or [[superficial]] BCC component overlying an [[invasive]] front showing basal cell carcinoma and [[squamous cell carcinoma]] [[histologic]] feature | ||
| | | | ||
|} | |} | ||
| Line 159: | Line 166: | ||
{{Reflist|2}} | {{Reflist|2}} | ||
{{WikiDoc Help Menu}} | {{WikiDoc Help Menu}} | ||
{{WikiDoc Sources}} | {{WikiDoc Sources}} | ||
[[Category:Up-To-Date]] | |||
[[Category:Oncology]] | [[Category:Oncology]] | ||
[[Category:Medicine]] | [[Category:Medicine]] | ||
[[Category:Dermatology]] | [[Category:Dermatology]] | ||
[[Category:Surgery]] | [[Category:Surgery]] | ||
Latest revision as of 18:25, 4 April 2019
|
Basal cell carcinoma Microchapters |
|
Diagnosis |
|---|
|
Case Studies |
|
Basal cell carcinoma pathophysiology On the Web |
|
American Roentgen Ray Society Images of Basal cell carcinoma pathophysiology |
|
Risk calculators and risk factors for Basal cell carcinoma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1];Associate Editor(s)-in-Chief: Maneesha Nandimandalam, M.B.B.S.[2]
Overview
Basal cell carcinoma is one of the most common skin cancers. It is commonly known as rodent ulcer due to its distinct morphology characterized by pearly pink nodules with telangiectasias, rolled borders, and central crusting with or without an ulcerating lesion. The majority common cause for the development of the basal cell carcinoma involves radiation exposure and mutations that involve many genes including sonic hedgehog gene, PTCH1 gene, and other gain-of-function mutations which further depend on the subtypes such as nodular, superficial, Infundibulocystic, fibroepithelial, morpheaform, infiltrative, micronodular, and basosquamous basal cell carcinomas.
Pathophysiology
Pathogenesis
The exact pathogenesis of basal cell carcinoma is not completely understood
Genetics
The development of basal cell carcinoma is the result of multiple genetic mutations such as sonic hedgehog pathway mutations, and PTCH1 gene mutations
- A number of aberrations involving the sonic hedgehog signaling pathway(SHH) are noted.[1][2][3][4]
- This pathway is vital for the regulation of cell growth, and differentiation and loss of inhibition of this pathway is associated with development of basal cell cancer.
- The majority of mutations in sporadic basal cell carcinoma and basal cell nevus syndrome(BCNS) patients occur in PTCH1 gene, a protein that inhibits smoothened gene (SMO).
- The second most common mutation in sporadic basal cell carcinoma and basal cell nevus syndrome(BCNS) patients are gain-of-function mutations of the smoothened gene (SMO).
- Loss of PTCH1 results in the failure of Smoothened inhibition, subsequently leading to increases in GLI1 levels, changes in transcription, and subsequent tumorigenesis.
- Gain-of-function smoothened(SMO) mutations also leads to increased GLI1 levels and tumorigenesis
| Loss of PTCH1 | Gain of function SMO | ||||||||||||||||||||||||
| Lack of SMO inhibition | Activation of SMO-GLI signaling | ||||||||||||||||||||||||
| ↑GLI1 levels | |||||||||||||||||||||||||
| Changes in transcription | |||||||||||||||||||||||||
| Tumorigenesis | |||||||||||||||||||||||||

Other Genetic Changes:
- Point mutations in the TP53 gene, the tumor supressor gene are the second most common genetic alteration noticed in BCCs
- Some mutations in the CDKN2A locus and in ras gene family (H-ras, K-ras, and N-ras) are also identified in a smaller proportion of sporadic BCCs
Enviromental Exposure
- Basal cell carcinomas develop in the basal cell layer of the skin.[5]
- Cumulative DNA damage caused by chronic sunlight exposure results in DNA mutations that predispose to the development of basal cell carcinoma.
- While DNA repair eliminates most UV-induced damage, not all cross-links are excised, which eventually results in mutations.
- Apart from the mutagenesis, sunlight depresses the local immune system, possibly decreasing immune surveillance for new tumor cells.
Gross and microscopic pathology
- On gross and microscopic histopathological analysis the characteristic findings of basal cell carcinoma are described as below:
- Basal cell carcinoma pathological features mainly depend upon the subtype. The following table summarizes them:[6][7]
| Subtypes of BCC | Gross features | Microscopic features | ||
| Findings | Images | Findings | Images | |
| Nodular |
|
 |
 | |
| Superficial |
|
 |
|
 |
| Infundibulocystic |
|
|||
| Fibroepithelial |
|
|
||
| Morpheaform |
 |
|
 | |
| Infiltrative |
|
 |
|
|
| Micronodular |
|
|||
| Basosquamous |
|
|||
Video
{{#ev:youtube|JnJXrFnvOKs}}
References
- ↑ Mohan SV, Chang AL (2014). "Advanced Basal Cell Carcinoma: Epidemiology and Therapeutic Innovations". Curr Dermatol Rep. 3: 40–45. doi:10.1007/s13671-014-0069-y. PMC 3931971. PMID 24587976.
- ↑ Pellegrini C, Maturo MG, Di Nardo L, Ciciarelli V, Gutiérrez García-Rodrigo C, Fargnoli MC (November 2017). "Understanding the Molecular Genetics of Basal Cell Carcinoma". Int J Mol Sci. 18 (11). doi:10.3390/ijms18112485. PMC 5713451. PMID 29165358.
- ↑ Yunoki T, Tabuchi Y, Hirano T, Miwa S, Imura J, Hayashi A (November 2018). "Gene networks in basal cell carcinoma of the eyelid, analyzed using gene expression profiling". Oncol Lett. 16 (5): 6729–6734. doi:10.3892/ol.2018.9484. PMC 6202553. PMID 30405815.
- ↑ Marzuka AG, Book SE (June 2015). "Basal cell carcinoma: pathogenesis, epidemiology, clinical features, diagnosis, histopathology, and management". Yale J Biol Med. 88 (2): 167–79. PMC 4445438. PMID 26029015.
- ↑ Montagna E, Lopes OS (2017). "Molecular basis of basal cell carcinoma". An Bras Dermatol. 92 (4): 517–520. doi:10.1590/abd1806-4841.20176544. PMC 5595599. PMID 28954101.
- ↑ Cameron, Michael C.; Lee, Erica; Hibler, Brian P.; Barker, Christopher A.; Mori, Shoko; Cordova, Miguel; Nehal, Kishwer S.; Rossi, Anthony M. (2019). "Basal cell carcinoma". Journal of the American Academy of Dermatology. 80 (2): 303–317. doi:10.1016/j.jaad.2018.03.060. ISSN 0190-9622.
- ↑ Sehgal VN, Chatterjee K, Pandhi D, Khurana A (2014). "Basal cell carcinoma: pathophysiology". Skinmed. 12 (3): 176–81. PMID 25134314.