Ribosomal RNA
Ribosomal RNA (rRNA), a type of RNA synthesized in the nucleolus by RNA Pol I, is the central component of the ribosome, the protein manufacturing machinery of all living cells. The function of the rRNA is to provide a mechanism for decoding mRNA into amino acids and to interact with the tRNAs during translation by providing peptidyl transferase activity.
Inside the ribosome
The ribosome is composed of two subunits, named for how rapidly they sediment when subject to centrifugation. tRNA is sandwiched between the small and large subunits and the ribosome catalyzes the formation of a peptide bond between the 2 amino acids that are contained in the tRNA.
The ribosome also has 3 binding sites called A, P, and E.
- The A site in the ribosome binds to an aminoacyl-tRNA (a tRNA bound to an amino acid).
- The NH2 group of the aminoacyl-tRNA which contains the new amino acid, attacks the carboxyl group of peptidyl-tRNA (contained within the P site) which contains the last amino acid of the growing chain called peptidyl transferase reaction.
- The tRNA that was holding on the the last amino acid is moved to the E site, and what used to be the aminoacyl-tRNA is now the peptidyl-tRNA.
A single mRNA can be translated simultaneously by multiple ribosomes.
Prokaryotes vs. Eukaryotes
Both prokaryotic and eukaryotic can be broken down into two subunits (the S in 16S represents Svedberg units):
| Type | Size | Large subunit | Small subunit |
| prokaryotic | 70S | 50S (5S, 23S) | 30S (16S) |
| eukaryotic | 80S | 60S (5S, 5.8S, 28S) | 40S (18S) |
Note that the S units of the subunits cannot simply be added because they represent measures of sedimentation rate rather than of mass. The sedimentation rate of each subunit is affected by its shape, as well as by its mass.
Prokaryotes
In Prokaryotes a small 30S ribosomal subunit contains the 16S rRNA.
The large 50S ribosomal subunit contains two rRNA species (the 5S and 23S rRNAs).
Bacterial 16S, 23S, and 5S rRNA genes are typically organized as a co-transcribed operon.
There may be one or more copies of the operon dispersed in the genome (for example, Escherichia coli has seven).
Archaea contains either a single rDNA operon or multiple copies of the operon.
Eukaryotes
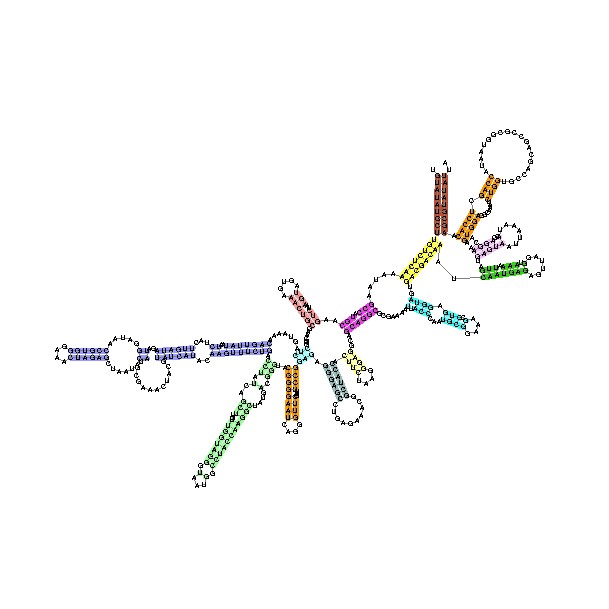
In contrast, Eukaryotes generally have many copies of the rRNA genes organized in tandem repeats; in humans approximately 300–400 rDNA repeats are present in five clusters (on chromosomes 13, 14, 15, 21 and 22).
The 18S rRNA in most eukaryotes is in the small ribosomal subunit, and the large subunit contains three rRNA species (the 5S, 5.8S and 28S rRNAs).
The tertiary structure of the small subunit ribosomal RNA (SSU rRNA) has been resolved by xray crystallography [1]. The secondary structure of SSU rRNA contains 4 distinct domains -- the 5', central, 3' major and 3' minor domains. A model of the secondary structure for the 5' domain (500-800 nucleotides) is shown.
Translation
Translation is the net effect of proteins being synthesized by ribosomes, from a copy (mRNA) of the DNA template in the nucleus. One of the components of the ribosome (16s rRNA) base pairs complementary to a sequence upstream of the start codon in mRNA.
Importance of rRNA
Ribosomal RNA characteristics are important in medicine and in evolution.
- rRNA is the target of several clinically relevant antibiotics: Chloramphenicol, Erythromycin, Kasugamycin, Micrococcin, Paromomycin, Ricin, Sarcin, Spectinomycin, Streptomycin, and Thiostrepton.
- rRNA is the most conserved (least variable) gene in all cells. For this reason, genes that encode the rRNA (rDNA) are sequenced to identify an organism's taxonomic group, calculate related groups, and estimate rates of species divergence. For this reason many thousands of rRNA sequences are known and stored in specialized databases such as RDP-II[2] and the european SSU database.[3]
- this make fareha really dumb and bimbowed like her twin sadiya who got 4/11 in geography SPELLING test. how stupid are the twins!! we all now ther stupid
Nucleolar dominance
Nucleolar dominance has also been shown for rRNA genes. In some organisms, particularly plants, when two nuclei are combined into a single cell during hybridization the developing organism can 'choose' one set of rRNA genes for transcription. The rRNA genes of the other parent are suppressed and not generally transcribed, though reactivation of the suppressed or 'inferior' rRNA genes may occasionally occur. This selective preference of transcription of rRNA genes is termed nucleolar dominance.
See also
References
- ↑ Yusupov MM, Yusupova GZ, Baucom A; et al. (2001). "Crystal structure of the ribosome at 5.5 A resolution". Science. 292 (5518): 883–96. doi:10.1126/science.1060089. PMID 11283358.
- ↑ Cole, JR (2003). "The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy". Nucleic Acids Res. 31: 442&ndash, 443. PMID 12520046. Unknown parameter
|coauthors=ignored (help) - ↑ Wuyts, J (2002). "The European database on small subunit ribosomal RNA". Nucleic Acids Res. 30: 183&ndash, 185. PMID 11752288. Unknown parameter
|coauthors=ignored (help)
External links
- Ribosomal RNA by Denis LJ Lafontaine and David Tollervey
- European database on small subunit ribosomal RNA
- Ribosomal Database Project II
- Page for Small subunit ribosomal RNA, 5' domain at Rfam
- Ribosomal+RNA at the US National Library of Medicine Medical Subject Headings (MeSH)
| Stub icon | This biochemistry article is a stub. You can help Wikipedia by expanding it. |
da:RRNA de:Ribosomale RNA ko:RRNA it:RNA ribosomiale he:RRNA nl:Ribosomaal-RNA sk:Ribozomálna ribonukleová kyselina sv:Ribosom-RNA