Ribosomal RNA: Difference between revisions
MishelhMagg (talk | contribs) No edit summary |
m (Robot: Automated text replacement (-{{SIB}} +, -{{EH}} +, -{{EJ}} +, -{{Editor Help}} +, -{{Editor Join}} +)) |
||
| (2 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
'''Ribosomal RNA''' (rRNA) | |||
==Overview== | |||
'''Ribosomal RNA''' ('''rRNA''') is the central component of the [[ribosome]], the protein manufacturing machinery of all living [[biological cell|cells]]. The function of the rRNA is to provide a mechanism for decoding [[mRNA]] into [[amino acid]]s and to interact with the [[tRNA]]s during [[Translation (biology)| translation]] by providing [[peptidyl transferase]] activity. | |||
==Inside the ribosome== | ==Inside the ribosome== | ||
| Line 7: | Line 10: | ||
* The A site in the ribosome binds to an aminoacyl-tRNA (a tRNA bound to an amino acid). | * The A site in the ribosome binds to an aminoacyl-tRNA (a tRNA bound to an amino acid). | ||
* The NH2 group of the aminoacyl-tRNA which contains the new amino acid, attacks the carboxyl group of peptidyl-tRNA (contained within the P site) which contains the last amino acid of the growing chain called [[peptidyl transferase reaction]]. | * The NH2 group of the aminoacyl-tRNA which contains the new amino acid, attacks the carboxyl group of peptidyl-tRNA (contained within the P site) which contains the last amino acid of the growing chain called [[peptidyl transferase reaction]]. | ||
* The tRNA that was holding on | * The tRNA that was holding on the last amino acid is moved to the E site, and what used to be the aminoacyl-tRNA is now the peptidyl-tRNA. | ||
A single mRNA can be translated simultaneously by multiple ribosomes. | A single mRNA can be translated simultaneously by multiple ribosomes. | ||
| Line 13: | Line 16: | ||
==Prokaryotes vs. Eukaryotes== | ==Prokaryotes vs. Eukaryotes== | ||
Both prokaryotic and eukaryotic can be broken down into two subunits (the S in 16S represents [[Svedberg]] units): | Both prokaryotic and eukaryotic can be broken down into two subunits (the S in [[16S ribosomal RNA|16S]] represents [[Svedberg]] units): | ||
{| class="wikitable" | {| class="wikitable" | ||
| '''Type''' || '''Size''' || '''Large subunit''' || '''Small subunit ''' | | '''Type''' || '''Size''' || '''Large subunit''' || '''Small subunit ''' | ||
|- | |- | ||
| prokaryotic || 70S || [[50S]] (5S, 23S) || [[30S]] (16S) | | prokaryotic || 70S || [[50S]] ([[5S ribosomal RNA|5S]], [[23S ribosomal RNA|23S]]) || [[30S]] ([[16S ribosomal RNA|16S]]) | ||
|- | |- | ||
| eukaryotic || 80S || 60S (5S, 5.8S, 28S) || 40S (18S) | | eukaryotic || 80S || [[60S]] ([[5S ribosomal RNA|5S]], [[5.8S ribosomal RNA|5.8S]], 28S) || 40S (18S) | ||
|} | |} | ||
| Line 26: | Line 29: | ||
===Prokaryotes=== | ===Prokaryotes=== | ||
In [[ | In [[prokaryotes]] a small 30S ribosomal subunit contains the [[16S ribosomal RNA|16S]] rRNA. | ||
The large 50S ribosomal subunit contains two rRNA species (the 5S and 23S rRNAs). | The large 50S ribosomal subunit contains two rRNA species (the 5S and [[23S ribosomal RNA|23S]] rRNAs). | ||
Bacterial 16S, 23S, and 5S rRNA genes are typically organized as a co-transcribed [[operon]]. | Bacterial [[16S ribosomal RNA|16S]], [[23S ribosomal RNA|23S]], and 5S rRNA genes are typically organized as a co-transcribed [[operon]]. | ||
There may be one or more copies of the operon dispersed in the [[genome]] (for example, [[Escherichia coli]] has seven). | There may be one or more copies of the [[operon]] dispersed in the [[genome]] (for example, ''[[Escherichia coli]]'' has seven). | ||
Archaea contains either a single rDNA operon or multiple copies of the operon. | [[Archaea]] contains either a single rDNA [[operon]] or multiple copies of the operon. | ||
The 3' end of the [[16S ribosomal RNA|16S]] rRNA (in a ribosome) binds to a sequence on the 5' end of mRNA called the [[Shine-Dalgarno sequence]]. | |||
===Eukaryotes=== | ===Eukaryotes=== | ||
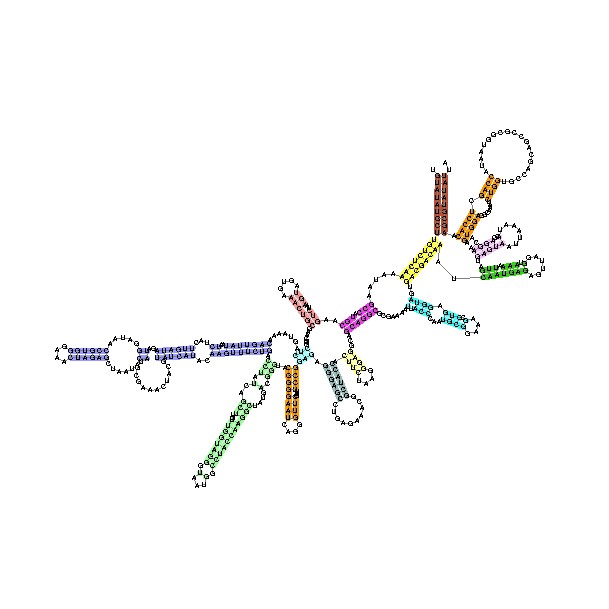
[[Image:RF00177.jpg|thumb|right|120px|Small subunit ribosomal RNA, 5' domain taken from the [[Rfam]] database. This example is [http:// | [[Image:RF00177.jpg|thumb|right|120px|Small subunit ribosomal RNA, 5' domain taken from the [[Rfam]] database. This example is [http://www.sanger.ac.uk/cgi-bin/Rfam/getacc?RF00177 RF00177] ]] | ||
In contrast, [[ | In contrast, [[eukaryotes]] generally have many copies of the rRNA genes organized in tandem repeats; in humans approximately 300–400 rDNA repeats are present in five clusters (on [[chromosomes]] 13, 14, 15, 21 and 22). | ||
The 18S rRNA in most eukaryotes is in the small ribosomal subunit, and the large subunit contains three rRNA species (the 5S, 5.8S and 28S rRNAs). | The [[18S ribosomal RNA|18S]] rRNA in most eukaryotes is in the small ribosomal subunit, and the large subunit contains three rRNA species (the [[5S ribosomal RNA|5S]], [[5.8S ribosomal RNA|5.8S]] and [[28S ribosomal RNA|28S]] rRNAs). | ||
The tertiary structure of the small subunit ribosomal RNA (SSU rRNA) has been resolved by | Mammalian cells have 2 mitochondrial ([[12S ribosomal RNA|12S]] and 16S) rRNA molecules and 4 types of cytoplasmic rRNA (28S, 5.8S, 5S (large ribosome subunit) and 18S (small subunit)). 28S, 5.8S, and 18S rRNAs are encoded by a single transcription unit (45S) separated by 2 Internally transcribed spacer (ITS). The 45S rDNA organized into 5 clusters (each has 30-40 repeats) on chromosomes 13, 14, 15, 21, and 22. These are transcribed by RNA polymerase I. 5S occurs in [[Tandemly arrayed genes|tandem arrays]] (~200-300 true 5S genes and many dispersed pseudogenes), the largest one on the chromosome 1q41-42. 5S rRNA is transcribed by RNA polymerase III. | ||
The tertiary structure of the small subunit ribosomal RNA (SSU rRNA) has been resolved by X-ray crystallography <ref name="pmid11283358">{{cite journal | author = Yusupov MM, Yusupova GZ, Baucom A, ''et al'' | title = Crystal structure of the ribosome at 5.5 A resolution | journal = Science | volume = 292 | issue = 5518 | pages = 883-96 | year = 2001 | pmid = 11283358 | doi = 10.1126/science.1060089 | issn = }}</ref>. The secondary structure of SSU rRNA contains 4 distinct domains -- the 5', central, 3' major and 3' minor domains. A model of the secondary structure for the 5' domain (500-800 nucleotides) is shown. | |||
==Translation== | ==Translation== | ||
[[Translation]] is the net effect of proteins being synthesized by ribosomes, from a copy (mRNA) of the DNA template in the nucleus. One of the components of the ribosome (16s rRNA) base pairs complementary to a sequence upstream of the [[start codon]] in mRNA. | [[Translation_(biology)|Translation]] is the net effect of proteins being synthesized by ribosomes, from a copy (mRNA) of the DNA template in the nucleus. One of the components of the ribosome (16s rRNA) base pairs complementary to a sequence upstream of the [[start codon]] in mRNA. | ||
==Importance of rRNA== | ==Importance of rRNA== | ||
Ribosomal RNA characteristics are important in [[medicine]] and in [[evolution]]. | Ribosomal RNA characteristics are important in [[medicine]] and in [[evolution]]. | ||
* rRNA is the target of several clinically relevant [[antibiotics]]: Chloramphenicol, Erythromycin, Kasugamycin, Micrococcin, Paromomycin, [[Ricin]], Sarcin, Spectinomycin, Streptomycin, and Thiostrepton. | * rRNA is the target of several clinically relevant [[antibiotics]]: [[Chloramphenicol]], [[Erythromycin]], [[Kasugamycin]], [[Micrococcin]], [[Paromomycin]], [[Ricin]], [[Sarcin]], [[Spectinomycin]], [[Streptomycin]], and [[Thiostrepton]]. | ||
* rRNA is the most conserved (least variable) gene in all cells. For this reason, genes that encode the rRNA (rDNA) are sequenced to identify an organism's [[taxonomic]] group, calculate related groups, and estimate rates of species divergence. For this reason many thousands of rRNA sequences are known and stored in specialized databases such as RDP-II<ref>{{cite journal | last = Cole | first = JR | coauthors = Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM | year = 2003 | title = The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy | journal = Nucleic Acids Res | volume = 31 | pages = 442–443 | id = PMID 12520046}}</ref> and the | * rRNA is the most conserved (least variable) gene in all cells. For this reason, genes that encode the rRNA (rDNA) are sequenced to identify an organism's [[taxonomic]] group, calculate related groups, and estimate rates of species divergence. For this reason many thousands of rRNA sequences are known and stored in specialized databases such as RDP-II<ref>{{cite journal | last = Cole | first = JR | coauthors = Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM | year = 2003 | title = The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy | journal = Nucleic Acids Res | volume = 31 | pages = 442–443 | id = PMID 12520046}}</ref> and the European SSU database.<ref>{{cite journal | last = Wuyts | first = J | coauthors = Van de Peer Y, Winkelmans T, De Wachter R | year = 2002 | title = The European database on small subunit ribosomal RNA | journal = Nucleic Acids Res | volume = 30 | pages = 183–185 | id = PMID 11752288}}</ref> | ||
==See also== | ==See also== | ||
| Line 67: | Line 70: | ||
==External links== | ==External links== | ||
* [http:// | * [http://www.his.se/upload/21749/Ribosomal%20RNA.pdf ''Ribosomal RNA''] by Denis LJ Lafontaine and David Tollervey | ||
* [http:// | * [http://www.psb.ugent.be/rRNA/ European database on small subunit ribosomal RNA] | ||
* [http:// | * [http://www.arb-silva.de SILVA Database Project] (also includes Eukaryotes and 23S / 28S) | ||
* [http://rdp.cme.msu.edu/ Ribosomal Database Project II] | |||
* [http://wiki.biomine.skelleftea.se/wiki/index.php/16S_ribosomal_RNA 16S rRNA, BioMineWiki] | |||
* {{Rfam|id=RF00177|name=Small subunit ribosomal RNA, 5' domain}} | * {{Rfam|id=RF00177|name=Small subunit ribosomal RNA, 5' domain}} | ||
* {{MeshName|Ribosomal+RNA}} | * {{MeshName|Ribosomal+RNA}} | ||
[[Category:Protein biosynthesis]] | [[Category:Protein biosynthesis]] | ||
[[Category:RNA]] | [[Category:RNA]] | ||
| Line 82: | Line 87: | ||
{{Nucleic acids}} | {{Nucleic acids}} | ||
[[ar:رنا الريبوسومي]] | |||
[[br:Trenkenn ribonukleek ribosom]] | |||
[[ca:ARN ribosòmic]] | |||
[[cs:RRNA]] | |||
[[da:RRNA]] | [[da:RRNA]] | ||
[[de:Ribosomale RNA]] | [[de:Ribosomale RNA]] | ||
[[el:Ριβοσωμικό RNA]] | |||
[[es:ARN ribosómico]] | [[es:ARN ribosómico]] | ||
[[fr:Acide ribonucléique ribosomique]] | [[fr:Acide ribonucléique ribosomique]] | ||
| Line 89: | Line 99: | ||
[[it:RNA ribosomiale]] | [[it:RNA ribosomiale]] | ||
[[he:RRNA]] | [[he:RRNA]] | ||
[[nl:Ribosomaal | [[la:RRNA]] | ||
[[nl:Ribosomaal RNA]] | |||
[[ja:リボソームRNA]] | [[ja:リボソームRNA]] | ||
[[pl:RRNA]] | [[pl:RRNA]] | ||
| Line 96: | Line 107: | ||
[[sk:Ribozomálna ribonukleová kyselina]] | [[sk:Ribozomálna ribonukleová kyselina]] | ||
[[sv:Ribosom-RNA]] | [[sv:Ribosom-RNA]] | ||
[[tr: | [[tr:Ribozomal RNA]] | ||
[[zh:核糖體RNA]] | [[zh:核糖體RNA]] | ||
{{WH}} | |||
{{WS}} | |||
{{jb1}} | |||
Latest revision as of 15:43, 20 August 2012
Overview
Ribosomal RNA (rRNA) is the central component of the ribosome, the protein manufacturing machinery of all living cells. The function of the rRNA is to provide a mechanism for decoding mRNA into amino acids and to interact with the tRNAs during translation by providing peptidyl transferase activity.
Inside the ribosome
The ribosome is composed of two subunits, named for how rapidly they sediment when subject to centrifugation. tRNA is sandwiched between the small and large subunits and the ribosome catalyzes the formation of a peptide bond between the 2 amino acids that are contained in the tRNA.
The ribosome also has 3 binding sites called A, P, and E.
- The A site in the ribosome binds to an aminoacyl-tRNA (a tRNA bound to an amino acid).
- The NH2 group of the aminoacyl-tRNA which contains the new amino acid, attacks the carboxyl group of peptidyl-tRNA (contained within the P site) which contains the last amino acid of the growing chain called peptidyl transferase reaction.
- The tRNA that was holding on the last amino acid is moved to the E site, and what used to be the aminoacyl-tRNA is now the peptidyl-tRNA.
A single mRNA can be translated simultaneously by multiple ribosomes.
Prokaryotes vs. Eukaryotes
Both prokaryotic and eukaryotic can be broken down into two subunits (the S in 16S represents Svedberg units):
| Type | Size | Large subunit | Small subunit |
| prokaryotic | 70S | 50S (5S, 23S) | 30S (16S) |
| eukaryotic | 80S | 60S (5S, 5.8S, 28S) | 40S (18S) |
Note that the S units of the subunits cannot simply be added because they represent measures of sedimentation rate rather than of mass. The sedimentation rate of each subunit is affected by its shape, as well as by its mass.
Prokaryotes
In prokaryotes a small 30S ribosomal subunit contains the 16S rRNA.
The large 50S ribosomal subunit contains two rRNA species (the 5S and 23S rRNAs).
Bacterial 16S, 23S, and 5S rRNA genes are typically organized as a co-transcribed operon.
There may be one or more copies of the operon dispersed in the genome (for example, Escherichia coli has seven).
Archaea contains either a single rDNA operon or multiple copies of the operon.
The 3' end of the 16S rRNA (in a ribosome) binds to a sequence on the 5' end of mRNA called the Shine-Dalgarno sequence.
Eukaryotes
In contrast, eukaryotes generally have many copies of the rRNA genes organized in tandem repeats; in humans approximately 300–400 rDNA repeats are present in five clusters (on chromosomes 13, 14, 15, 21 and 22).
The 18S rRNA in most eukaryotes is in the small ribosomal subunit, and the large subunit contains three rRNA species (the 5S, 5.8S and 28S rRNAs).
Mammalian cells have 2 mitochondrial (12S and 16S) rRNA molecules and 4 types of cytoplasmic rRNA (28S, 5.8S, 5S (large ribosome subunit) and 18S (small subunit)). 28S, 5.8S, and 18S rRNAs are encoded by a single transcription unit (45S) separated by 2 Internally transcribed spacer (ITS). The 45S rDNA organized into 5 clusters (each has 30-40 repeats) on chromosomes 13, 14, 15, 21, and 22. These are transcribed by RNA polymerase I. 5S occurs in tandem arrays (~200-300 true 5S genes and many dispersed pseudogenes), the largest one on the chromosome 1q41-42. 5S rRNA is transcribed by RNA polymerase III.
The tertiary structure of the small subunit ribosomal RNA (SSU rRNA) has been resolved by X-ray crystallography [1]. The secondary structure of SSU rRNA contains 4 distinct domains -- the 5', central, 3' major and 3' minor domains. A model of the secondary structure for the 5' domain (500-800 nucleotides) is shown.
Translation
Translation is the net effect of proteins being synthesized by ribosomes, from a copy (mRNA) of the DNA template in the nucleus. One of the components of the ribosome (16s rRNA) base pairs complementary to a sequence upstream of the start codon in mRNA.
Importance of rRNA
Ribosomal RNA characteristics are important in medicine and in evolution.
- rRNA is the target of several clinically relevant antibiotics: Chloramphenicol, Erythromycin, Kasugamycin, Micrococcin, Paromomycin, Ricin, Sarcin, Spectinomycin, Streptomycin, and Thiostrepton.
- rRNA is the most conserved (least variable) gene in all cells. For this reason, genes that encode the rRNA (rDNA) are sequenced to identify an organism's taxonomic group, calculate related groups, and estimate rates of species divergence. For this reason many thousands of rRNA sequences are known and stored in specialized databases such as RDP-II[2] and the European SSU database.[3]
See also
References
- ↑ Yusupov MM, Yusupova GZ, Baucom A; et al. (2001). "Crystal structure of the ribosome at 5.5 A resolution". Science. 292 (5518): 883–96. doi:10.1126/science.1060089. PMID 11283358.
- ↑ Cole, JR (2003). "The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy". Nucleic Acids Res. 31: 442&ndash, 443. PMID 12520046. Unknown parameter
|coauthors=ignored (help) - ↑ Wuyts, J (2002). "The European database on small subunit ribosomal RNA". Nucleic Acids Res. 30: 183&ndash, 185. PMID 11752288. Unknown parameter
|coauthors=ignored (help)
External links
- Ribosomal RNA by Denis LJ Lafontaine and David Tollervey
- European database on small subunit ribosomal RNA
- SILVA Database Project (also includes Eukaryotes and 23S / 28S)
- Ribosomal Database Project II
- 16S rRNA, BioMineWiki
- Page for Small subunit ribosomal RNA, 5' domain at Rfam
- Ribosomal+RNA at the US National Library of Medicine Medical Subject Headings (MeSH)
ar:رنا الريبوسومي br:Trenkenn ribonukleek ribosom ca:ARN ribosòmic cs:RRNA da:RRNA de:Ribosomale RNA el:Ριβοσωμικό RNA ko:RRNA it:RNA ribosomiale he:RRNA la:RRNA nl:Ribosomaal RNA sk:Ribozomálna ribonukleová kyselina sv:Ribosom-RNA