Epithelioid sarcoma pathophysiology: Difference between revisions
| Line 19: | Line 19: | ||
==Genetics== | ==Genetics== | ||
* The most common genetic mutation (found in 80-90% of epithelioid sarcomas) is the inactivation of the [[SMARCB1]] gene, or the loss of INI-1 function, which is thought to be a major contributor to disease progression.<ref name="pmid19033866">{{cite journal |last1=Hornick |first1=Jason L. |last2=Dal Cin |first2=Paola |last3=Fletcher |first3=Christopher D.M. |title=Loss of INI1 Expression is Characteristic of Both Conventional and Proximal-type Epithelioid Sarcoma |journal=The American Journal of Surgical Pathology |volume=33 |issue=4 |pages=542–50 |year=2009 |pmid=19033866 |doi=10.1097/PAS.0b013e3181882c54 }}</ref><ref name="pmid15899790">{{cite journal |last1=Modena |first1=Piergiorgio |last2=Lualdi |first2=Elena |last3=Facchinetti |first3=Federica |last4=Galli |first4=Lisa |last5=Teixeira |first5=Manuel R. |last6=Pilotti |first6=Silvana |last7=Sozzi |first7=Gabriella |title=''SMARCB1/INI1'' Tumor Suppressor Gene Is Frequently Inactivated in Epithelioid Sarcomas |journal=Cancer Research |volume=65 |issue=10 |pages=4012–9 |year=2005 |pmid=15899790 |doi=10.1158/0008-5472.CAN-04-3050 }}</ref> Epithelioid sarcoma typically contains chromosome 22q11.2 mutations or deletions and 8q gains, particularly i(8) (>q10). Aberrations of 18q and 8q, as well as recurrent gains at 11q13, have also been observed.<ref>{{cite journal |last1=Lushnikova |first1=Tamara |last2=Knuutila |first2=Sakari |last3=Miettinen |first3=Markku |title=DNA Copy Number Changes in Epithelioid Sarcoma and Its Variants: A Comparative Genomic Hybridization Study |journal=Modern Pathology |volume=13 |issue=10 |pages=1092–6 |year=2000 |pmid=11048803 |doi=10.1038/modpathol.3880203 }}</ref><ref>{{cite journal |last1=Nishio |first1=Jun |last2=Iwasaki |first2=Hiroshi |last3=Nabeshima |first3=Kazuki |last4=Ishiguro |first4=Masako |last5=Naumann |first5=Sabine |last6=Isayama |first6=Teruto |last7=Naito |first7=Masatoshi |last8=Kaneko |first8=Yasuhiko |last9=Kikuchi |first9=Masahiro |last10=Bridge |first10=Julia |title=Establishment of a new human epithelioid sarcoma cell line, FU-EPS-1: Molecular cytogenetic characterization by use of spectral karyotyping and comparative genomic hybridization |journal=International Journal of Oncology |volume=27 |issue=2 |pages=361–9 |year=2005 |pmid=16010416 |doi=10.3892/ijo.27.2.361 }}</ref><ref name="pmid15578074">{{cite journal |last1=Lin |first1=Lin |last2=Hicks |first2=David |last3=Xu |first3=Bo |last4=Sigel |first4=Jessica E |last5=Bergfeld |first5=Wilma F |last6=Montgomery |first6=Elizabeth |last7=Fisher |first7=Cyril |last8=Hartke |first8=Marybeth |last9=Tubbs |first9=Raymond |last10=Goldblum |first10=John R |title=Expression profile and molecular genetic regulation of cyclin D1 expression in epithelioid sarcoma |journal=Modern Pathology |volume=18 |issue=5 |pages=705–9 |year=2005 |pmid=15578074 |doi=10.1038/modpathol.3800349 }}</ref> | * The most common genetic mutation (found in 80-90% of epithelioid sarcomas) is the inactivation of the [[SMARCB1]] gene, or the loss of INI-1 function, which is thought to be a major contributor to disease progression.<ref name="pmid19033866">{{cite journal |last1=Hornick |first1=Jason L. |last2=Dal Cin |first2=Paola |last3=Fletcher |first3=Christopher D.M. |title=Loss of INI1 Expression is Characteristic of Both Conventional and Proximal-type Epithelioid Sarcoma |journal=The American Journal of Surgical Pathology |volume=33 |issue=4 |pages=542–50 |year=2009 |pmid=19033866 |doi=10.1097/PAS.0b013e3181882c54 }}</ref><ref name="pmid15899790">{{cite journal |last1=Modena |first1=Piergiorgio |last2=Lualdi |first2=Elena |last3=Facchinetti |first3=Federica |last4=Galli |first4=Lisa |last5=Teixeira |first5=Manuel R. |last6=Pilotti |first6=Silvana |last7=Sozzi |first7=Gabriella |title=''SMARCB1/INI1'' Tumor Suppressor Gene Is Frequently Inactivated in Epithelioid Sarcomas |journal=Cancer Research |volume=65 |issue=10 |pages=4012–9 |year=2005 |pmid=15899790 |doi=10.1158/0008-5472.CAN-04-3050 }}</ref> Epithelioid sarcoma typically contains chromosome 22q11.2 mutations or deletions and 8q gains, particularly i(8) (>q10). Aberrations of 18q and 8q, as well as recurrent gains at 11q13, have also been observed.<ref>{{cite journal |last1=Lushnikova |first1=Tamara |last2=Knuutila |first2=Sakari |last3=Miettinen |first3=Markku |title=DNA Copy Number Changes in Epithelioid Sarcoma and Its Variants: A Comparative Genomic Hybridization Study |journal=Modern Pathology |volume=13 |issue=10 |pages=1092–6 |year=2000 |pmid=11048803 |doi=10.1038/modpathol.3880203 }}</ref><ref>{{cite journal |last1=Nishio |first1=Jun |last2=Iwasaki |first2=Hiroshi |last3=Nabeshima |first3=Kazuki |last4=Ishiguro |first4=Masako |last5=Naumann |first5=Sabine |last6=Isayama |first6=Teruto |last7=Naito |first7=Masatoshi |last8=Kaneko |first8=Yasuhiko |last9=Kikuchi |first9=Masahiro |last10=Bridge |first10=Julia |title=Establishment of a new human epithelioid sarcoma cell line, FU-EPS-1: Molecular cytogenetic characterization by use of spectral karyotyping and comparative genomic hybridization |journal=International Journal of Oncology |volume=27 |issue=2 |pages=361–9 |year=2005 |pmid=16010416 |doi=10.3892/ijo.27.2.361 }}</ref><ref name="pmid15578074">{{cite journal |last1=Lin |first1=Lin |last2=Hicks |first2=David |last3=Xu |first3=Bo |last4=Sigel |first4=Jessica E |last5=Bergfeld |first5=Wilma F |last6=Montgomery |first6=Elizabeth |last7=Fisher |first7=Cyril |last8=Hartke |first8=Marybeth |last9=Tubbs |first9=Raymond |last10=Goldblum |first10=John R |title=Expression profile and molecular genetic regulation of cyclin D1 expression in epithelioid sarcoma |journal=Modern Pathology |volume=18 |issue=5 |pages=705–9 |year=2005 |pmid=15578074 |doi=10.1038/modpathol.3800349 }}</ref> | ||
* The ''SMARCB1'' gene (also termed BAF47, INI1, or hSNF5) is located on chromosome [[22q11.2]]<ref name="pmid19033866" /> and codes for a member of the [[SWI/SNF]] chromatin remodeling complex. Loss of SMARCB1 function is the most common genetic mutation observed in epithelioid sarcoma, and this dysfunction is likely a major driver of disease progression. SMARCB1 is a core protein subunit of the 15 subunit SWI/SNF (or BAF) complex involved in regulating the nucleosome architecture of our genome<ref name="pmid19033866" /> and has been shown to be a potent [[tumor suppressor gene]],<ref>{{cite journal |last1=Kahali |first1=Bhaskar |last2=Yu |first2=Jinlong |last3=Marquez |first3=Stefanie B. |last4=Thompson |first4=Kenneth W. |last5=Liang |first5=Shermi Y. |last6=Lu |first6=Li |last7=Reisman |first7=David |title=The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors |journal=Oncotarget |volume=5 |issue=10 |pages=3316–32 |year=2014 |pmid=24913006 |pmc=4102812 |doi=10.18632/oncotarget.1945 }}</ref><ref name="pmid15899790" /> | * The ''SMARCB1'' gene (also termed BAF47, INI1, or hSNF5) is located on chromosome [[22q11.2]]<ref name="pmid19033866" /> and codes for a member of the [[SWI/SNF]] chromatin remodeling complex. Loss of SMARCB1 function is the most common genetic mutation observed in epithelioid sarcoma, and this dysfunction is likely a major driver of disease progression. SMARCB1 is a core protein subunit of the 15 subunit SWI/SNF (or BAF) complex involved in regulating the nucleosome architecture of our genome<ref name="pmid19033866" /> and has been shown to be a potent [[tumor suppressor gene]], meaning that its primary role is to control cell division and to even halt division under appropriate circumstances (i.e. signals to over-replicate).<ref>{{cite journal |last1=Kahali |first1=Bhaskar |last2=Yu |first2=Jinlong |last3=Marquez |first3=Stefanie B. |last4=Thompson |first4=Kenneth W. |last5=Liang |first5=Shermi Y. |last6=Lu |first6=Li |last7=Reisman |first7=David |title=The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors |journal=Oncotarget |volume=5 |issue=10 |pages=3316–32 |year=2014 |pmid=24913006 |pmc=4102812 |doi=10.18632/oncotarget.1945 }}</ref><ref name="pmid15899790" /> As this tumor suppressor is commonly inactivated in epithelioid sarcoma, cell division can fail to appropriately halt, resulting in unregulated cellular growth and the formation of cancer tumors. Several research teams are currently developing techniques to reverse this loss of genetic function characteristic of epithelioid sarcoma.<ref name="LevDina" /> | ||
==Molecular Biology== | ==Molecular Biology== | ||
Revision as of 14:44, 29 February 2016
|
Epithelioid sarcoma Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Epithelioid sarcoma pathophysiology On the Web |
|
American Roentgen Ray Society Images of Epithelioid sarcoma pathophysiology |
|
Risk calculators and risk factors for Epithelioid sarcoma pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]; Associate Editor(s)-in-Chief: Ammu Susheela, M.D. [2]
Overview
Mutations in the SMARCB1 gene cause epithelioid sarcoma. On gross pathology, a solid, multinodular mass with a glistening gray tan appearance and multiple areas of hemorrhage and necrosis are characteristic findings of epithelioid sarcoma. On microscopic histopathological analysis, central necrosis surrounded by bland, polygonal cells with eosinophilic cytoplasm and peripheral spindling, desmoplasia, focal calcification, or metaplastic ossification are characteristic findings of epithelioid sarcoma.
Pathogenesis
- Epithelioid sarcoma is the second most common soft tissue sarcoma in hand. Epithelioid sarcoma is also the sixth most common soft tissue sarcoma in the upper extremity.
- Primary site of epithelioid sarcoma is upper distal extremities. Other rare sites of epithelioid sarcoma are:
- Epithelioid sarcoma is a rare soft tissue sarcoma arising from mesenchymal tissue and characterized by epithelioid-like features.
- Epithelioid sarcoma accounts for less than 1% of all soft tissue sarcomas. Epithelioid sarcoma commonly presents itself in the distal limbs (fingers, hands, forearms, or feet) of young adults as a small, soft mass or a series of bumps. A proximal version has also been described, frequently occurring in the upper extremities.[1] Rare cases have been reported in the pelvis, vulva, penis, and spine.
- Epithelioid sarcoma most commonly strikes young adults, yet no age group is immune. The disease has a tendency to develop local recurrences and metastasis thereafter to regional lymph nodes, lung, bone, brain, and other locations, including the scalp.[2] Generally speaking, epithelioid sarcoma has a high rate of relapse after initial treatment and tends to recur locally (at or near the original tumor site). Epithelioid sarcoma also demonstrates lymphatic spread (in 22-48% of cases), and metastasis (in 21-63% of cases).[3] These events, as well as advanced stage (progression) and grade (aggressiveness), are predictive of an overall worse outcome. The overall five-year survival rate for epithelioid sarcoma is anywhere from 25 to 78%. Importantly, the 10-year and 15-year survival rate drops off significantly.[4][5] Associated with a more positive outcome are younger age, female vs. male sex, distal vs. proximal location, smaller tumor size, and negative margins upon tumor resection.[6][5]
Genetics
- SMARCB1 gene is involved in the pathogenesis of epithelioid sarcoma.
Genetics
- The most common genetic mutation (found in 80-90% of epithelioid sarcomas) is the inactivation of the SMARCB1 gene, or the loss of INI-1 function, which is thought to be a major contributor to disease progression.[7][8] Epithelioid sarcoma typically contains chromosome 22q11.2 mutations or deletions and 8q gains, particularly i(8) (>q10). Aberrations of 18q and 8q, as well as recurrent gains at 11q13, have also been observed.[9][10][11]
- The SMARCB1 gene (also termed BAF47, INI1, or hSNF5) is located on chromosome 22q11.2[7] and codes for a member of the SWI/SNF chromatin remodeling complex. Loss of SMARCB1 function is the most common genetic mutation observed in epithelioid sarcoma, and this dysfunction is likely a major driver of disease progression. SMARCB1 is a core protein subunit of the 15 subunit SWI/SNF (or BAF) complex involved in regulating the nucleosome architecture of our genome[7] and has been shown to be a potent tumor suppressor gene, meaning that its primary role is to control cell division and to even halt division under appropriate circumstances (i.e. signals to over-replicate).[12][8] As this tumor suppressor is commonly inactivated in epithelioid sarcoma, cell division can fail to appropriately halt, resulting in unregulated cellular growth and the formation of cancer tumors. Several research teams are currently developing techniques to reverse this loss of genetic function characteristic of epithelioid sarcoma.[4]
Molecular Biology
VEGF
VEGF (vascular endothelial growth factor) is often over-expressed in epithelioid sarcoma.[13] This is a critical pathway in angiogenesis, a process that cancer cells use to form new blood vessels, which provide necessary elements to the tumor for tumor survival. Anti-VEGF agents such as pazopanib have shown promise across several different carcinomas and in soft tissue sarcomas.[14] In one case study, a patient with advanced metastatic vulvar epithelioid sarcoma showed a partial resolution of both lung and pleural metastases when pazopanib was administered, whereas all other therapies had failed[15]
MET
MET (mesenchymal to epithelial transition) is another biological pathway that is likely involved in the development and progression of epithelioid sarcoma.[16][17] c-MET is a tyrosine kinase oncogene, and its signaling pathway has been implicated in a variety of malignancies, including many cancers.
Sonic hedgehog and Notch
The Sonic hedgehog and Notch signaling pathways are also suspected to be up-regulated in epithelioid sarcoma. These cell signaling pathways control cellular proliferation and differentiation. They are also involved in cancer stem cell coordination and disease invasiveness and metastasis. Hhat inhibitors (such as RU-SKI 43) block the Sonic hedgehog signaling pathway by inhibiting hedgehog palmitoyl acytl-transferase. Current trials are investigating Notch inhibitors against epithelioid sarcoma.[18]
mTOR
The frequent hyperactivation of mTOR (mammalian target of rapamycin) signaling has also been observed in epithelioid sarcoma.[17][19] The mTOR pathway has been described as a “master switch” for cellular catabolism and anabolism, and it can enhance cell cycle progression, cell survival, and block normal cell death (apoptosis).[14] Interestingly, it has been demonstrated that simply blocking mTOR signaling can result in the reactivation of the AKT pathway, negating much of the anti-mTOR’s efficacy.[17] This reactivation of AKT has been shown to be c-MET-dependent,[17] resulting in the rationale that blocking both mTOR and c-MET concurrently would show increased efficacy.
EGFR
The over-expression of epidermal growth factor receptor (EGFR) has been reported in a majority of epithelioid sarcomas.[19][20] EGFR is a member of the HER receptor family. Upon ligand binding, EGFR phosphorylation triggers the activation of downstream signaling pathways involved in critical cellular functions such as proliferation, survival, and angiogenesis.[21] In-vitro and in-vivo laboratory experiments have demonstrated that the blockade of EGFR in epithelioid sarcoma results in decreased cell proliferation, increased apoptosis, and abrogated invasion and migration capacities.[19] Of interest, while the simple blockade of EGFR with a single agent has shown limited results in the clinical setting, when used as part of a combination regime (where an EGFR inhibitor is combined with an mTOR inhibitor), a synergism has been observed, and superior tumor growth inhibition has been demonstrated.[19]
CD109
CD109 is often expressed in advanced epithelioid sarcoma and is thought to mark the cancer stem cell (or cancer initiating cell) of the disease.[22] Its level of expression has also been shown to be predictive of outcome. Cancer stem cells are a small population of tumor cells characterized by general chemo-resistance, the ability to self-renew, multi-differentiation potential, dormancy capabilities, and tumorigenesis. Therefore, cancer stem cells are thought to play key roles in the progression and relapse of cancer.
Cyclin D1
Cyclin D1 is a protein requisite for cell cycle progression and has been shown to be up-regulated in epithelioid sarcoma.[11] Cyclin D-1 is a regulator of cyclin-dependent kinases (CDK4 and CDK6). It has been shown to interact with the retinoblastoma protein (a tumor suppressor gene), CDK4 and CDK6, thyroid hormone receptor beta, and nuclear receptor coactivator 1, among others.[11] Cyclin D and CDKs promote cell cycle progression by releasing transcription factors that are important for the initiation of DNA replication. Abnormal levels of cyclin D-1 may promote rapid cell division in epithelioid sarcoma.
Gross Pathology
- Gross pathological features are:
- Solid, multinodular mass
- Glistening gray tan appearance
- Multiple areas of hemorrhage and necrosis
Microscopic Pathology
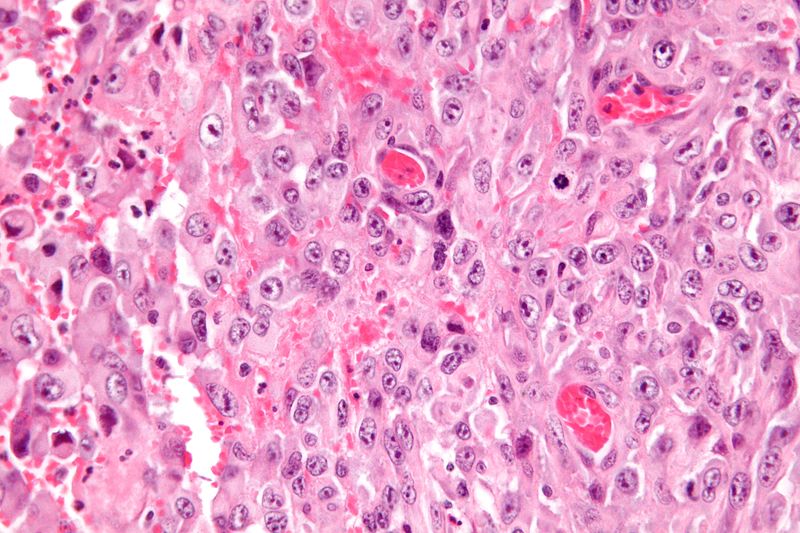
- Histologically, epithelioid sarcoma forms nodules with central necrosis surrounded by bland, polygonal cells with eosinophilic cytoplasm and peripheral spindling.[2] Epithelioid sarcomas typically express vimentin, cytokeratins, epithelial membrane antigen, and CD34, whereas they are usually negative for S100, desmin, and FLI-1.[2] They typically stain positive for CA125.[23]
- Desmoplasia, focal calcification, or metaplastic ossification
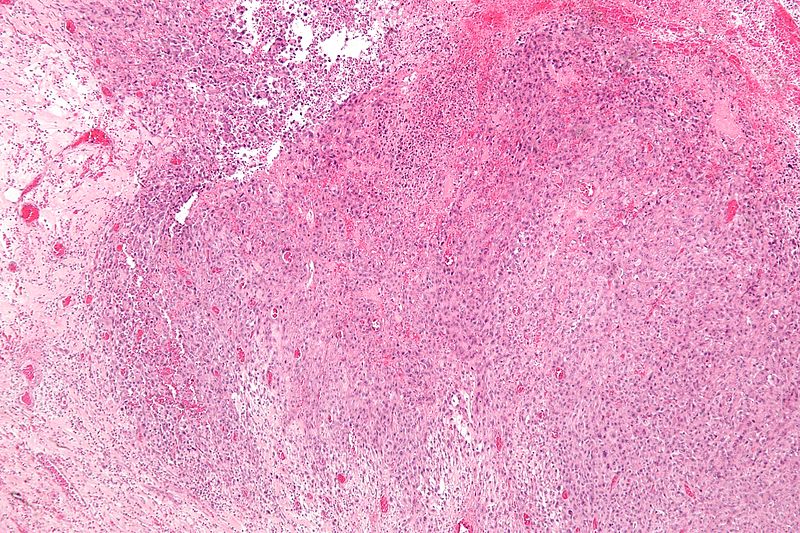
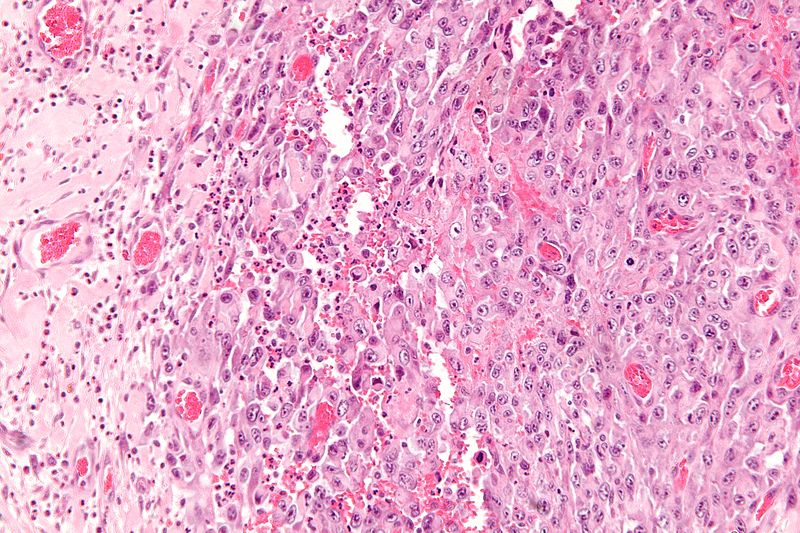
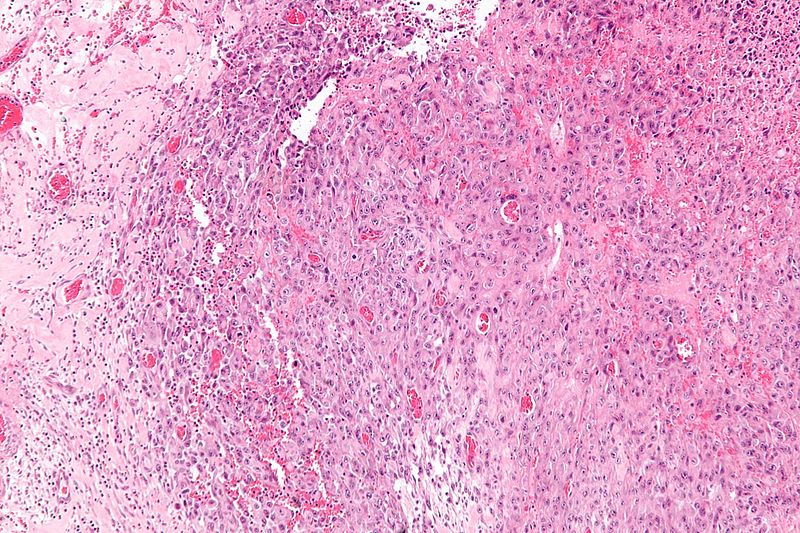
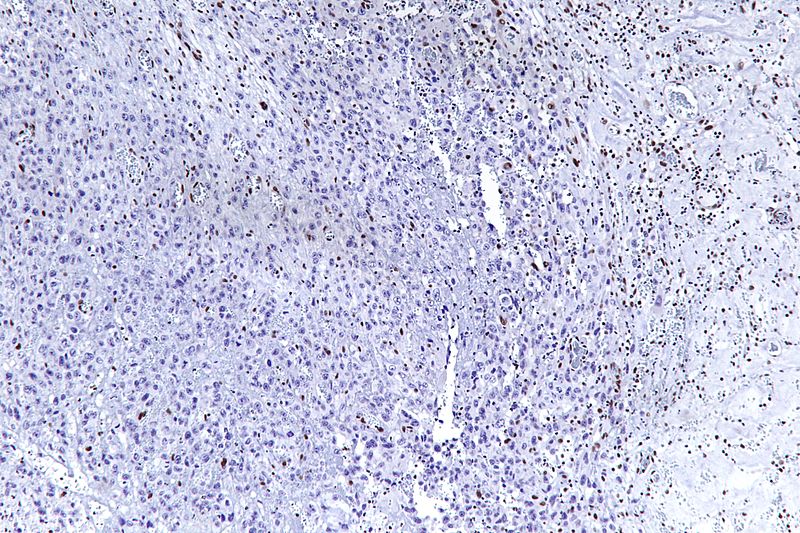
Gallery
-
Low magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
-
Low magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
-
Intermediate magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
-
High magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
-
Very high magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
-
Very high magnification micrograph of epithelioid sarcoma<ref> Epithelioid sarcoma librepathology
References
- ↑ Guillou, L; Wadden, C; Coindre, JM; Krausz, T; Fletcher, CD (1997). "'Proximal-type' epithelioid sarcoma, a distinctive aggressive neoplasm showing rhabdoid features. Clinicopathologic, immunohistochemical, and ultrastructural study of a series". The American Journal of Surgical Pathology. 21 (2): 130–46. doi:10.1097/00000478-199702000-00002. PMID 9042279.
- ↑ 2.0 2.1 2.2 Armah, Henry B. Armah; Parwani, Anil V. (2009). "Epithelioid sarcoma". Archives of Pathology & Laboratory Medicine. 133 (5): 814–9. doi:10.1043/1543-2165-133.5.814 (inactive October 6, 2015). PMID 19415960.
- ↑ Levy, Antonin; Le Péchoux, Cécile; Terrier, Philippe; Bouaita, Ryan; Domont, Julien; Mir, Olivier; Coppola, Sarah; Honoré, Charles; Le Cesne, Axel; Bonvalot, Sylvie (2014). "Epithelioid Sarcoma: Need for a Multimodal Approach to Maximize the Chances of Curative Conservative Treatment". Annals of Surgical Oncology. 21 (1): 269–76. doi:10.1245/s10434-013-3247-4. PMID 24046109.
- ↑ 4.0 4.1 Lev, Dina. "Epigenetic reprogramming of epitheliold sarcoma: a role for INI1-HDAC crosstalk".[dead link]
- ↑ 5.0 5.1 Casanova, Michela; Ferrari, Andrea; Collini, Paola; Bisogno, Gianni; Alaggio, Rita; Cecchetto, Giovanni; Gronchi, Alessandro; Meazza, Cristina; Garaventa, Alberto; Di Cataldo, Andrea; Carli, Modesto (2006). "Epithelioid sarcoma in children and adolescents". Cancer. 106 (3): 708–17. doi:10.1002/cncr.21630. PMID 16353216.
- ↑ Jawad, Muhammad Umar; Extein, Jason; Min, Elijah S.; Scully, Sean P. (2009). "Prognostic Factors for Survival in Patients with Epithelioid Sarcoma: 441 Cases from the SEER Database". Clinical Orthopaedics and Related Research. 467 (11): 2939–48. doi:10.1007/s11999-009-0749-2. PMC 2758965. PMID 19224301.
- ↑ 7.0 7.1 7.2 Hornick, Jason L.; Dal Cin, Paola; Fletcher, Christopher D.M. (2009). "Loss of INI1 Expression is Characteristic of Both Conventional and Proximal-type Epithelioid Sarcoma". The American Journal of Surgical Pathology. 33 (4): 542–50. doi:10.1097/PAS.0b013e3181882c54. PMID 19033866.
- ↑ 8.0 8.1 Modena, Piergiorgio; Lualdi, Elena; Facchinetti, Federica; Galli, Lisa; Teixeira, Manuel R.; Pilotti, Silvana; Sozzi, Gabriella (2005). "SMARCB1/INI1 Tumor Suppressor Gene Is Frequently Inactivated in Epithelioid Sarcomas". Cancer Research. 65 (10): 4012–9. doi:10.1158/0008-5472.CAN-04-3050. PMID 15899790.
- ↑ Lushnikova, Tamara; Knuutila, Sakari; Miettinen, Markku (2000). "DNA Copy Number Changes in Epithelioid Sarcoma and Its Variants: A Comparative Genomic Hybridization Study". Modern Pathology. 13 (10): 1092–6. doi:10.1038/modpathol.3880203. PMID 11048803.
- ↑ Nishio, Jun; Iwasaki, Hiroshi; Nabeshima, Kazuki; Ishiguro, Masako; Naumann, Sabine; Isayama, Teruto; Naito, Masatoshi; Kaneko, Yasuhiko; Kikuchi, Masahiro; Bridge, Julia (2005). "Establishment of a new human epithelioid sarcoma cell line, FU-EPS-1: Molecular cytogenetic characterization by use of spectral karyotyping and comparative genomic hybridization". International Journal of Oncology. 27 (2): 361–9. doi:10.3892/ijo.27.2.361. PMID 16010416.
- ↑ 11.0 11.1 11.2 Lin, Lin; Hicks, David; Xu, Bo; Sigel, Jessica E; Bergfeld, Wilma F; Montgomery, Elizabeth; Fisher, Cyril; Hartke, Marybeth; Tubbs, Raymond; Goldblum, John R (2005). "Expression profile and molecular genetic regulation of cyclin D1 expression in epithelioid sarcoma". Modern Pathology. 18 (5): 705–9. doi:10.1038/modpathol.3800349. PMID 15578074.
- ↑ Kahali, Bhaskar; Yu, Jinlong; Marquez, Stefanie B.; Thompson, Kenneth W.; Liang, Shermi Y.; Lu, Li; Reisman, David (2014). "The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors". Oncotarget. 5 (10): 3316–32. doi:10.18632/oncotarget.1945. PMC 4102812. PMID 24913006.
- ↑ Kuhnen, Cornelius; Lehnhardt, Marcus; Tolnay, Edina; Muehlberger, Thomas; Vogt, Peter M.; Müller, Klaus-Michael (2000). "Patterns of expression and secretion of vascular endothelial growth factor in malignant soft-tissue tumours". Journal of Cancer Research and Clinical Oncology. 126 (4): 219–25. doi:10.1007/s004320050036. PMID 10782895.
- ↑ 14.0 14.1 Martín Liberal, Juan; Lagares-Tena, Laura; Sáinz-Jaspeado, Miguel; Mateo-Lozano, Silvia; García del Muro, Xavier; Tirado, Oscar M. (2012). "Targeted Therapies in Sarcomas: Challenging the Challenge". Sarcoma. 2012: 626094. doi:10.1155/2012/626094. PMC 3372278. PMID 22701332.
- ↑ Chung, Hye Won. "The treatment of pazopanib on vulvar epithelioid sarcoma: A case report and review of literature".[unreliable medical source?]
- ↑ Kuhnen, C.; Tolnay, Edina; Steinau, Hans Ulrich; Voss, Bruno; Müller, Klaus-Michael (1998). "Expression of c-Met receptor and hepatocyte growth factor/scatter factor in synovial sarcoma and epithelioid sarcoma". Virchows Archiv. 432 (4): 337–42. doi:10.1007/s004280050175. PMID 9565343.
- ↑ 17.0 17.1 17.2 17.3 Imura, Yoshinori; Yasui, Hirohiko; Outani, Hidetatsu; Wakamatsu, Toru; Hamada, Kenichiro; Nakai, Takaaki; Yamada, Shutaro; Myoui, Akira; Araki, Nobuhito; Ueda, Takafumi; Itoh, Kazuyuki; Yoshikawa, Hideki; Naka, Norifumi (2014). "Combined targeting of mTOR and c-MET signaling pathways for effective management of epithelioid sarcoma". Molecular Cancer. 13: 185. doi:10.1186/1476-4598-13-185. PMC 4249599. PMID 25098767.
- ↑ Clinical trial number NCT01154452 for "Vismodegib and Gamma-Secretase/Notch Signalling Pathway Inhibitor RO4929097 in Treating Patients With Advanced or Metastatic Sarcoma" at ClinicalTrials.gov
- ↑ 19.0 19.1 19.2 19.3 Xie, X.; Ghadimi, M. P. H.; Young, E. D.; Belousov, R.; Zhu, Q.-s.; Liu, J.; Lopez, G.; Colombo, C.; Peng, T.; Reynoso, D.; Hornick, J. L.; Lazar, A. J.; Lev, D. (2011). "Combining EGFR and mTOR Blockade for the Treatment of Epithelioid Sarcoma". Clinical Cancer Research. 17 (18): 5901–12. doi:10.1158/1078-0432.CCR-11-0660. PMC 3176924. PMID 21821699.
- ↑ Cascio, Michael J; O'Donnell, Richard J; Horvai, Andrew E (2010). "Epithelioid sarcoma expresses epidermal growth factor receptor but gene amplification and kinase domain mutations are rare". Modern Pathology. 23 (4): 574–80. doi:10.1038/modpathol.2010.2. PMID 20118913.
- ↑ Yang, J.-L.; Hannan, M.T.; Russell, P.J.; Crowe, P.J. (2006). "Expression of HER1/EGFR protein in human soft tissue sarcomas". European Journal of Surgical Oncology. 32 (4): 466–8. doi:10.1016/j.ejso.2006.01.012. PMID 16524687.
- ↑ Ahmad, Aamir; Emori, Makoto; Tsukahara, Tomohide; Murase, Masaki; Kano, Masanobu; Murata, Kenji; Takahashi, Akari; Kubo, Terufumi; Asanuma, Hiroko; Yasuda, Kazuyo; Kochin, Vitaly; Kaya, Mitsunori; Nagoya, Satoshi; Nishio, Jun; Iwasaki, Hiroshi; Sonoda, Tomoko; Hasegawa, Tadashi; Torigoe, Toshihiko; Wada, Takuro; Yamashita, Toshihiko; Sato, Noriyuki (2013). "High Expression of CD109 Antigen Regulates the Phenotype of Cancer Stem-Like Cells/Cancer-Initiating Cells in the Novel Epithelioid Sarcoma Cell Line ESX and Is Related to Poor Prognosis of Soft Tissue Sarcoma". PLoS ONE. 8 (12): e84187. doi:10.1371/journal.pone.0084187. PMC 3869840. PMID 24376795.
- ↑ Kato, Hiroshi; Hatori, Masahito; Kokubun, Shoichi; Watanabe, Mika; Smith, Richard A; Hotta, Tetsuo; Ogose, Akira; Morita, Tetsuro; Murakami, Takashi; Aiba, Setsuya (2004). "CA125 expression in epithelioid sarcoma". Japanese Journal of Clinical Oncology. 34 (3): 149–54. doi:10.1093/jjco/hyh027. PMID 15078911.