RuBisCO
Ribulose-1,5-bisphosphate carboxylase/oxygenase, most commonly known by the shorter name RuBisCO [1] , is an enzyme (EC 4.1.1.39) that is used in the Calvin cycle to catalyze the first major step of carbon fixation, a process by which the atoms of atmospheric carbon dioxide are made available to organisms in the form of energy-rich molecules such as sucrose. RuBisCO catalyzes either the carboxylation or oxygenation of ribulose-1,5-bisphosphate (also known as RuBP) with carbon dioxide or oxygen.
RuBisCO is very important in terms of biological impact because it catalyzes the most commonly used chemical reaction by which inorganic carbon enters the biosphere. RuBisCO is apparently the most abundant protein in leaves, and it may be the most abundant protein on Earth[2]. Given its important role in the biosphere, there are currently efforts to genetically engineer crop plants so as to contain more efficient RuBisCO (see below).
Structure
In plants, algae, cyanobacteria, and phototropic and chemoautotropic proteobacteria the enzyme usually consists of two types of protein subunit, called the large chain (L, about 55,000 Da) and the small chain (S, about 13,000 Da)[3]. The enzymatically active substrate (ribulose 1,5-bisphosphate) binding sites are located in the large chains that form dimers as shown in Figure 1 (above, right) in which amino acids from each large chain contribute to the binding sites. A total of eight large chain dimers and eight small chains assemble into a larger complex of about 540,000 Da[4]. In some proteobacteria and dinoflagellates, enzymes consisting of only large subunits have been found [5].
Magnesium ions (Mg2+) are needed for enzymatic activity. Correct positioning of Mg2+ in the active site of the enzyme involves addition of an "activating" carbon dioxide molecule (CO2) to a lysine in the active site (forming a carbamate)[6]. Formation of the carbamate is favored by an alkaline pH. The pH and the concentration of magnesium ions in the fluid compartment (in plants, the stroma of the chloroplast[7]) increases in the light. The role of changing pH and magnesium ion levels in the regulation of RuBisCO enzyme activity is discussed below.
Enzymatic activity
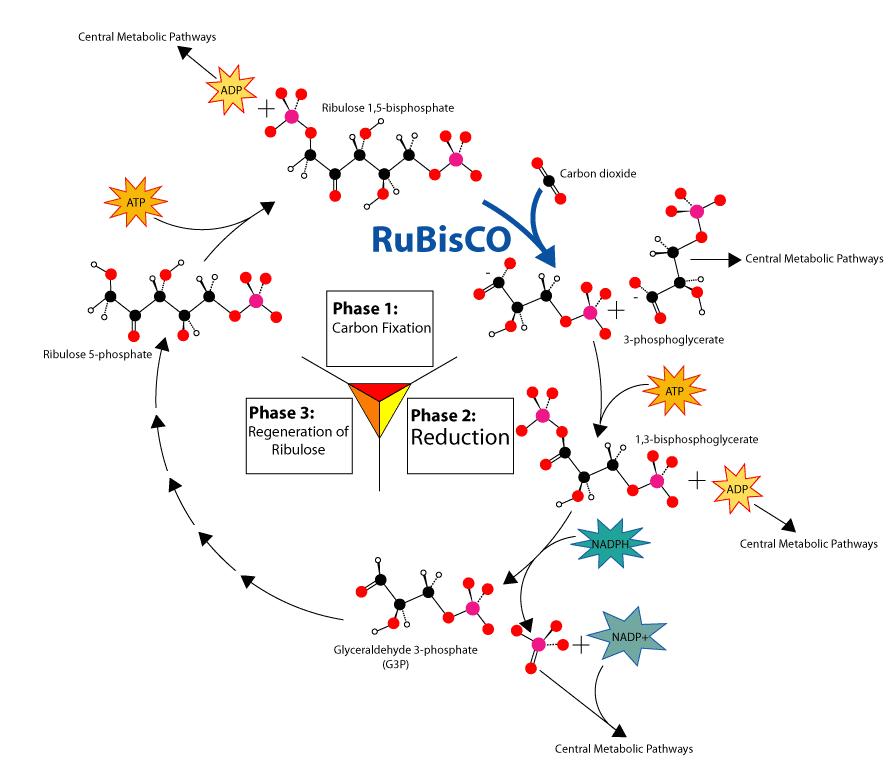
As shown in Figure 2 (left), RuBisCO is one of many enzymes in the Calvin cycle.
Substrates. During carbon fixation, the substrate molecules for RuBisCO are ribulose 1,5-bisphosphate, carbon dioxide (distinct from the "activating" carbon dioxide) and water [8]. RuBisCO can also allow a reaction to occur with molecular oxygen (O2) instead of carbon dioxide (CO2).
Products. When carbon dioxide is the substrate, the product of the carboxylase reaction is a highly unstable six-carbon phosphorylated intermediate which virtually instantaneously decays into two molecules of glycerate 3-phosphate . The extremely unstable actual molecule created by the initial carboxylation was unknown until 1988 when it was isolated. The 3-phosphoglycerate can be used to produce larger molecules such as glucose. When molecular oxygen is the substrate, the products of the oxygenase reaction are phosphoglycolate and 3-phosphoglycerate. Phosphoglycolate initiates a sequence of reactions called photorespiration which involves enzymes and cytochromes located in the mitochondria and peroxisomes. In this process, two molecules of phosphoglycolate are converted to one molecule of carbon dioxide and one molecule of 3-phosphoglycerate, which can reenter the Calvin cycle. Some of the phosphoglycolate entering this pathway can be retained by plants to produce other molecules such as glycine. At air levels of carbon dioxide and oxygen, the ratio of the reactions is about 4 to 1, which results in a net carbon dioxide fixation of only 3.5. Thus the inability of the enzyme to prevent the reaction with oxygen greatly reduces the photosynthetic potential of many plants. Some plants, many algae and photosynthetic bacteria have overcome this limitation by devising means to increase the concentration of carbon dioxide around the enzyme, including C4 carbon fixation, crassulacean acid metabolism and using pyrenoid.
Rate of enzymatic activity. Some enzymes typically can carry out thousands of chemical reactions each second. However, RuBisCO is slow, being able to "fix" only 3 carbon dioxide molecules each second. Nevertheless, because of its extremely large concentration, under most conditions, and when light is not otherwise limiting photosynthesis, the reaction of RuBisCO responds positively to increasing carbon dioxide concentration, therefore the concentration of carbon dioxide is limiting. The ultimate rate-limiting factor of the Calvin cycle is RuBisCo that cannot be ameliorated in short time by any other factor [1].
Regulation of its enzymatic activity
RuBisCO is usually only active during the day because ribulose 1,5-bisphosphate is not being produced in the dark, due to the regulation of several other enzymes in the Calvin cycle. In addition, the activity of Rubisco is coordinated with that of the other enzymes of the Calvin cycle in several ways:
- Regulation by ions. Upon illumination of the chloroplasts, the pH of the stroma rises from 7.0 to 8.0 because of the proton (hydrogen ion, H+) gradient created across the thylakoid membrane[9]. At the same time, magnesium ions (Mg2+) move out of the thylakoids, increasing the concentration of magnesium in the stroma of the chloroplasts. RuBisCO has a high optimal pH (can be >9.0, depending on the magnesium ion concentration) and thus becomes "activated" by the addition of carbon dioxide and magnesium to the active sites as described above.
- Regulation by activase. In plants and some algae, another enzyme, RuBisCO activase[10] is required to allow the rapid formation of the critical carbamate in the active site of RuBisCO[11]. Activase is required because the ribulose 1,5-bisphosphate (RuBP) substrate binds more strongly to the active sites lacking the carbamate and markedly slows down the "activation" process. In the light, RuBisCO activase promotes the release of the inhibitory, or in some views storage [2], RuBP from the catalytic sites. Activase is also required in some plants (e.g. tobacco and many beans) because in darkness, RuBisCO [3] is inhibited by a competitive inhibitor synthesized by these plants, a substrate analog 2-Carboxy-D-arabitinol 1-phosphate (CA1P)[12]. CA1P binds tightly to the active site of carbamylated RuBisCO and inhibits catalytic activity. In the light, RuBisCO activase also promotes the release of CA1P from the catalytic sites. After the CA1P is released from RuBisCO, it is rapidly converted to a non-inhibitory form by a light-activated CA1P-phosphatase. Finally, once every several hundred reactions, the normal reactions with carbon dioxide or oxygen are not completed and other inhibitory substrate analogs are formed in the active site. Once again, RuBisCO activase can promote the release of these analogs from the catalytic sites and maintain the enzyme in a catalytically active form. In the initial reaction of RuBisCO in the light, the RuBP that was separated from RuBisCO binds with the carbamylated enzyme and after proton abstraction produces Enediol that can react with carbon dioxide. A limitation of either RuBisCO or RuBP at any stage will make the reaction insensitive to any other factor including carbon dioxide. For this reason models that are based on a limitation of RuBisCO at low carbon dioxide levels such as compensation point, cannot support life on the planet[4].
The properties of activase limit the photosynthetic potential of plants at high temperatures [13]. CA1P has also been shown to keep Rubisco in a conformation that is protected from proteolysis[14].
- Regulation by ATP/ADP and stromal reduction/oxidation state through the activase. The removal of the inhibitory RuBP, CA1P, and the other inhibitory substrate analogs by activase requires the consumption of ATP. This reaction is inhibited by the presence of ADP and thus activase activity depends on the ratio of these compounds in the chloroplast stroma. Furthermore in most plants, the sensitivity of activase to the ratio of ATP/ADP is modified by the stromal reduction/oxidation (redox) state through another small regulatory protein, thioredoxin. In this manner, the activity of activase and the activation state of Rubisco can be modulated in response to light intensity and thus the rate of formation of the ribulose 1,5-bisphosphate substrate[15].
- Regulation by phosphate. In cyanobacteria, inorganic phosphate (Pi) participates in the co-ordinated regulation of photosynthesis. Pi binds to the RuBisCO active site and to another site on the large chain where it can influence transitions between activated and less active conformations of the enzyme. Activation of bacterial RuBisCO might be particularly sensitive to Pi levels which can act in the same way as RuBisCO activase in higher plants [16].
- Regulation by carbon dioxide. Since carbon dioxide and oxygen compete at the active site of RuBisCO, carbon fixation by RuBisCO can be enhanced by increasing the carbon dioxide level in the compartment containing RuBisCO (chloroplast stroma). Several times during the evolution of plants, mechanisms have evolved for increasing the level of carbon dioxide in the stroma (see C4 carbon fixation). The use of oxygen as a substrate is an apparently-puzzling process, since it seems to throw away captured energy. However it may be a mechanism for preventing overload during periods of high light flux. This weakness in the enzyme is the cause of photorespiration such that healthy leaves in bright light may have zero net carbon fixation when the ratio of O2 to CO2 reaches a threshold at which oxygen is fixed instead of carbon. This phenomenon is primarily temperature dependent. High temperature decreases the concentration of CO2 dissolved in the moisture in the leaf tissues. This phenomenon is also related to water stress. Since plant leaves are evaporatively cooled, limited water causes high leaf temperatures. C4 plants use the enzyme PEP carboxylase initially, which has a higher affinity for CO2. The process first makes a 4-carbon intermediate compound which is shuttled into a site of C3 photosynthesis then de-carboxylated releasing CO2 to boost the concentration of CO2, hence the name C4 plants.
Crassulacean acid metabolism (CAM) plants keep their stomata (on the underside of the leaf) closed during the day, which conserves water but prevents photosynthesis, which requires CO2 to pass by gas exchange through these openings. Evaporation through the upper side of a leaf is prevented by a layer of wax.
Genetic engineering
Since RuBisCO is often rate limiting for photosynthesis in plants, it may be possible to improve photosynthetic efficiency by modifying RuBisCO genes in plants to increase its catalytic activity and/or decrease the rate of the oxygenation activity[17]. Approaches that have begun to be investigated include expressing RuBisCO genes from one organism in another organism, increasing the level of expression of RuBisCO subunits, expressing RuBisCO small chains from the chloroplast DNA, and altering RuBisCO genes so as to try to increase specificity for carbon dioxide or otherwise increase the rate of carbon fixation[18].
One particularly interesting avenue is to introduce RuBisCO variants with naturally high specificity values such as the ones from the red alga Galdieria partita into plants. This would be expected to improve the photosynthetic efficiency of crop plants [19]. Important advances in this area include the replacement of the tobacco enzyme with that of the purple photosynthetic bacterium Rhodospirillum rubrum [20].
A recent theory [21] explores the trade off between the relative specificity (i.e. ability to favour CO2 fixation over O2 incorporation, which leads to the energetically wasteful process of photorespiration) and the rate at which product is formed. The authors conclude that RuBisCO may actually have evolved to reach a point of 'near perfection'[5] in many plants (with widely varying substrate availabilities and environmental conditions), reaching a compromise between specificity and rate of reaction.
RuBisCO and carbon sequestration
In view of greenhouse gas-induced climate change, it is speculated whether forests could be fertilized with nitrogen to increase foliar biomass in forests and thereby raise RuBisCO concentration to increase carbon sequestration and fixation in tree cellulosic structures (e.g., trunks, branches).[22][23]
References
- ↑ The term Rubisco was, in 1979, coined humorously by David Eisenberg at a seminar honouring the retirement of the early, prominent rubisco researcher, Sam Wildman. The abbreviation was derived from the full name (Ribulose-1,5-bisphosphate carboxylase/oxygenase) and also alluded to the snack food trade name "Nabisco" in reference to Wildman's attempts to create edible tobacco leaves. Wildman SG (2002) Along the trail from Faction I protein to Rubisco (ribulose bisphosphate carboxylase-oxygenase). Photosynth Res 73:243–250; Archie R. Portis Jr. & Martin A. J. Parry (2007) Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective Photosynth Res 94:121–143
- ↑ The Cell—A Molecular Approach. 2nd ed. by Geoffrey M. Cooper, published by Sinauer Associates, Inc. (2000) Sunderland (MA). Online textbook. The Cooper text suggests that RuBisCO is the most abundant protein on Earth (Chapter 10, The Chloroplast Genome). A recent article by Dhingra et al, suggests that RuBisCO accounts for 30–50% of total soluble protein in chloroplasts (full text article online: Template:Entrez Pubmed).
- ↑ The large chain gene is part of the chloroplast DNA molecule in plants (Entrez GeneID: ). There are typically several related small chain genes in the nucleus of plant cells and the small chains are imported to the stromal compartment of chloroplasts from the cytosol by crossing the outer chloroplast membrane (full text article online: Template:Entrez Pubmed). Arabidopsis thaliana has four RuBisCO small chain genes (see: Template:Entrez Pubmed). The pattern of how large chains and small chains assemble is illustrated in Figure 3 (right).
- ↑ Biochemistry by Jeremy M. Berg, John L. Tymoczko, and Lubert Stryer. Published by W. H. Freeman and Co. (2002) New York. Online textbook. Figure 20 in the Stryer textbook shows a color-coded ribbon diagram of the structural components of eukaryotic RuBisCO. Figure 1 (on this page, near top) shows another view of the structure.
- ↑ The structure of RuBisCO from the photosynthetic bacterium Rhodospirillum rubrum has been determined by X-ray crystallography, see: Template:Protein Data Bank. A comparison of the structures of eukaryotic and bacterial RuBisCO is shown in the Protein Data Bank feature article on Rubisco.
- ↑ Molecular Cell Biology, 4th edition, by Harvey Lodish, Arnold Berk, S. Lawrence Zipursky, Paul Matsudaira, David Baltimore and James E. Darnell. Published by W. H. Freeman & Co. (2000) New York. Online textbook. Figure 16-48 shows a structural model of the active site, including the involvement of magnesium. The Protein Data Bank feature article on RuBisCO also includes a model of magnesium at the active site.
- ↑ The Lodish textbook describes the localization of RuBisCO to the stromal space of chloroplasts. Figure 17-7 illustrates how RuBisCO small subunits move into the chloroplast stroma and assemble with the large subunits.
- ↑ The chemical reactions catalyzed by RuBisCO are described in the online Biochemistry textbook by Stryer et al.
- ↑ Figure 20.14 in the textbook by Stryer et al. illustrates the light-dependent movement of hydrogen and magnesium ions that are important for Light Regulation of the Calvin Cycle. The movement of protons into thylakoids is driven by light and is fundamental to ATP synthesis in chloroplasts.
- ↑ "Rubisco activase—Rubisco's catalytic chaperone." by A. R. Portis, Jr in Photosynthesis Research (2003), volume 75, pages 11–27. (see: Template:Entrez Pubmed).
- ↑ "Characteristics of photosynthesis in rice plants transformed with an antisense Rubisco activase gene" by S. H. Jin, D. A. Jiang, X. Q. Li and J. W. Sun. Transgenic plants that were genetically engineered to have reduced levels of RuBisCO activase were shown to have reduced photosynthesis (see: Template:Entrez Pubmed).
- ↑ "Incorporation of carbon from photosynthetic products into 2-carboxyarabinitol-1-phosphate and 2-carboxyarabinitol." by P. J. Andralojc, G. W. Dawson, M. A. Parry and A. J. Keys in Biochemical Journal (1994), volume 304, pages 781–6. (full text online: Template:Entrez Pubmed).
- ↑ "Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. by S. J. Crafts-Brandner and M. E. Salvucci in Proceedings of the National Academy of Science USA (2000), volume 97, pages 12937–8. (full text online:Template:Entrez Pubmed).
- ↑ "2'-carboxy-D-arabitinol 1-phosphate protects ribulose 1, 5-bisphosphate carboxylase/oxygenase against proteolytic breakdown" by S. Khan, P. J. Andralojc, P. J. Lea and M. A. Parry in European Journal of Biochemistry (1999), volume 266, pages 840–7. (full text online: Template:Entrez Pubmed).
- ↑ "Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform.." by N. Zhang, R. Kallis, R. G. Ewy, A. R. Portis Jr in Proceedings of the National Academy of Science USA (2002), volume 99, pages 3330–4 (full text online:Template:Entrez Pubmed).
- ↑ "Activation of cyanobacterial RuBP-carboxylase/oxygenase is facilitated by inorganic phosphate via two independent mechanisms." by Yehouda Marcus and Michael Gurevitz in European Journal of Biochemistry (2000), volume 267, pages 5995–6003. (full text online:Template:Entrez Pubmed).
- ↑ "Rubisco: structure, regulatory interactions, and possibilities for a better enzyme." by R. J Spreitzer and M. E. Salvucci in Annual Review of Plant Biology (2003) volume 53, page 449–75 (see: Template:Entrez Pubmed).
- ↑ "Manipulation of Rubisco: the amount, activity, function and regulation." by M. A. Parry, P. J. Andralojc, R. A. Mitchell, P. J. Madgwick and A. J. Keys in Journal of Experimental Botany (2003) volume 54, page 1321–33. (full text online: Template:Entrez Pubmed)
- ↑ "Whitney, S. M. and T. J. Andrews (2001). "Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco." Proceedings of the National Academy of Sciences of the United States of America 98(25):." (Template:Entrez Pubmed)
- ↑ "Andrews, T. J. and S. M. Whitney (2003). "Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants." Archives of Biochemistry and Biophysics 414(2): 159-169." (Template:Entrez Pubmed)
- ↑ G. G. Tcherkez, G. D. Farquhar and T. J. Andrews (2006). "Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized." Proceedings of the National Academy of Sciences of the United States of America 103(19):.Template:Entrez Pubmed
- ↑ P. Högberg (2007). "Nitrogen impacts on forest carbon." Nature 447: 781-782.
- ↑ F. Magnani, M. Mencuccini, M. Borghetti, P. Berbigier, F. Berninger, S. Delzon, A. Grelle, P. Hari, P.G. Jarvis, P. Kolari, A.S. Kowalski, H. Lankreijer, B.E. Law, A. Lindroth, D. Loustau, G. Manca, J.B. Moncrieff, M. Rayment, V. Tedeschi, B. Valentini, J. Grace (2007). "The human footprint in the carbon cycle of temperate and boreal forests." Nature 447: 848-850.
Further reading
- Sugawara H, Yamamoto H, Shibata N, Inoue T, Okada S, Miyake C, Yokota A, Kai Y. Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita. J Biol Chem 1999; 274:15655–61. Fulltext. PMID.
- Portis AR Jr, Parry MAJ. Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective. Photosynth Res 2007; 94:121–43. Template:Entrez Pubmed
See also
External links
- See here for the mechanism of the RuBisCO-catalysed reaction
- Rubisco: Protein Data Bank entry
- The Plant Kingdom's sloth: Protein spotlight article on the "slothful" enzyme RuBisCO
ca:RuBisCO cs:Rubisco da:Rubisco de:Rubisco eo:Rubisko id:Rubisco it:Ribulosio-bifosfato carbossilasi nl:Rubisco sv:Rubisco