RNA polymerase: Difference between revisions
Adeeljamil (talk | contribs) No edit summary |
Adeeljamil (talk | contribs) No edit summary |
||
| Line 4: | Line 4: | ||
{{EH}} | {{EH}} | ||
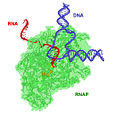
'''RNA polymerase''' ('''RNAP''' or '''RNApol''') is an [[enzyme]] that makes an [[RNA]] copy of a [[DNA]] or RNA template. In [[cell (biology)|cell]]s, RNAP is needed for constructing RNA chains from DNA [[gene]]s, a process called [[Transcription (genetics)|transcription]]. RNA polymerase enzymes are essential to life and are found in all organisms and many [[virus]]es. In chemical terms, RNAP is a [[nucleotidyl transferase]] that [[polymerization|polymerizes]] [[ribonucleotide]]s at the [[3']] | '''RNA polymerase''' ('''RNAP''' or '''RNApol''') is an [[enzyme]] that makes an [[RNA]] copy of a [[DNA]] or RNA template. In [[cell (biology)|cell]]s, RNAP is needed for constructing RNA chains from DNA [[gene]]s, a process called [[Transcription (genetics)|transcription]]. RNA polymerase enzymes are essential to life and are found in all organisms and many [[virus]]es. In chemical terms, RNAP is a [[nucleotidyl transferase]] that [[polymerization|polymerizes]] [[ribonucleotide]]s at the [[3' end]]of an RNA transcript. | ||
==History== | ==History== | ||
RNAP was discovered independently by Sam Weiss and Jerard Hurwitz in 1960.<ref>{{cite journal|author=Jerard Hurwitz|year=2005|month=Dec|title=The Discovery of RNA Polymerase|journal=Journal of Biological Chemistry |volume=280|issue=52|pages=42477-85|pmid=16230341|doi=10.1074/jbc.X500006200}}</ref> By this time the 1959 [[Nobel Prize]] in Medicine had been awarded to Severo Ochoa and Arthur Kornberg for the discovery of what was believed to be RNAP<ref>[http://nobelprize.org/nobel_prizes/medicine/laureates/1959/ Nobel Prize 1959]</ref>, but instead turned out to be a [[ribonuclease]]. | RNAP was discovered independently by Sam Weiss and Jerard Hurwitz in 1960.<ref>{{cite journal|author=Jerard Hurwitz|year=2005|month=Dec|title=The Discovery of RNA Polymerase|journal=Journal of Biological Chemistry |volume=280|issue=52|pages=42477-85|pmid=16230341|doi=10.1074/jbc.X500006200}}</ref> By this time the 1959 [[Nobel Prize]] in Medicine had been awarded to Severo Ochoa and Arthur Kornberg for the discovery of what was believed to be RNAP<ref>[http://nobelprize.org/nobel_prizes/medicine/laureates/1959/ Nobel Prize 1959]</ref>, but instead turned out to be a [[ribonuclease]]. | ||
The 2006 Nobel Prize in Chemistry was awarded to | The 2006 Nobel Prize in Chemistry was awarded to Roger Kornberg for creating detailed molecular images of RNA polymerase during various stages of the transcription process.<ref>[http://nobelprize.org/nobel_prizes/chemistry/laureates/2006/index.html Nobel Prize in Chemistry 2006]</ref> | ||
==Control of transcription== | ==Control of transcription== | ||
Revision as of 09:04, 23 April 2009
|
WikiDoc Resources for RNA polymerase |
|
Articles |
|---|
|
Most recent articles on RNA polymerase Most cited articles on RNA polymerase |
|
Media |
|
Powerpoint slides on RNA polymerase |
|
Evidence Based Medicine |
|
Clinical Trials |
|
Ongoing Trials on RNA polymerase at Clinical Trials.gov Trial results on RNA polymerase Clinical Trials on RNA polymerase at Google
|
|
Guidelines / Policies / Govt |
|
US National Guidelines Clearinghouse on RNA polymerase NICE Guidance on RNA polymerase
|
|
Books |
|
News |
|
Commentary |
|
Definitions |
|
Patient Resources / Community |
|
Patient resources on RNA polymerase Discussion groups on RNA polymerase Patient Handouts on RNA polymerase Directions to Hospitals Treating RNA polymerase Risk calculators and risk factors for RNA polymerase
|
|
Healthcare Provider Resources |
|
Causes & Risk Factors for RNA polymerase |
|
Continuing Medical Education (CME) |
|
International |
|
|
|
Business |
|
Experimental / Informatics |
Please Take Over This Page and Apply to be Editor-In-Chief for this topic: There can be one or more than one Editor-In-Chief. You may also apply to be an Associate Editor-In-Chief of one of the subtopics below. Please mail us [1] to indicate your interest in serving either as an Editor-In-Chief of the entire topic or as an Associate Editor-In-Chief for a subtopic. Please be sure to attach your CV and or biographical sketch.
RNA polymerase (RNAP or RNApol) is an enzyme that makes an RNA copy of a DNA or RNA template. In cells, RNAP is needed for constructing RNA chains from DNA genes, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses. In chemical terms, RNAP is a nucleotidyl transferase that polymerizes ribonucleotides at the 3' endof an RNA transcript.
History
RNAP was discovered independently by Sam Weiss and Jerard Hurwitz in 1960.[1] By this time the 1959 Nobel Prize in Medicine had been awarded to Severo Ochoa and Arthur Kornberg for the discovery of what was believed to be RNAP[2], but instead turned out to be a ribonuclease.
The 2006 Nobel Prize in Chemistry was awarded to Roger Kornberg for creating detailed molecular images of RNA polymerase during various stages of the transcription process.[3]
Control of transcription
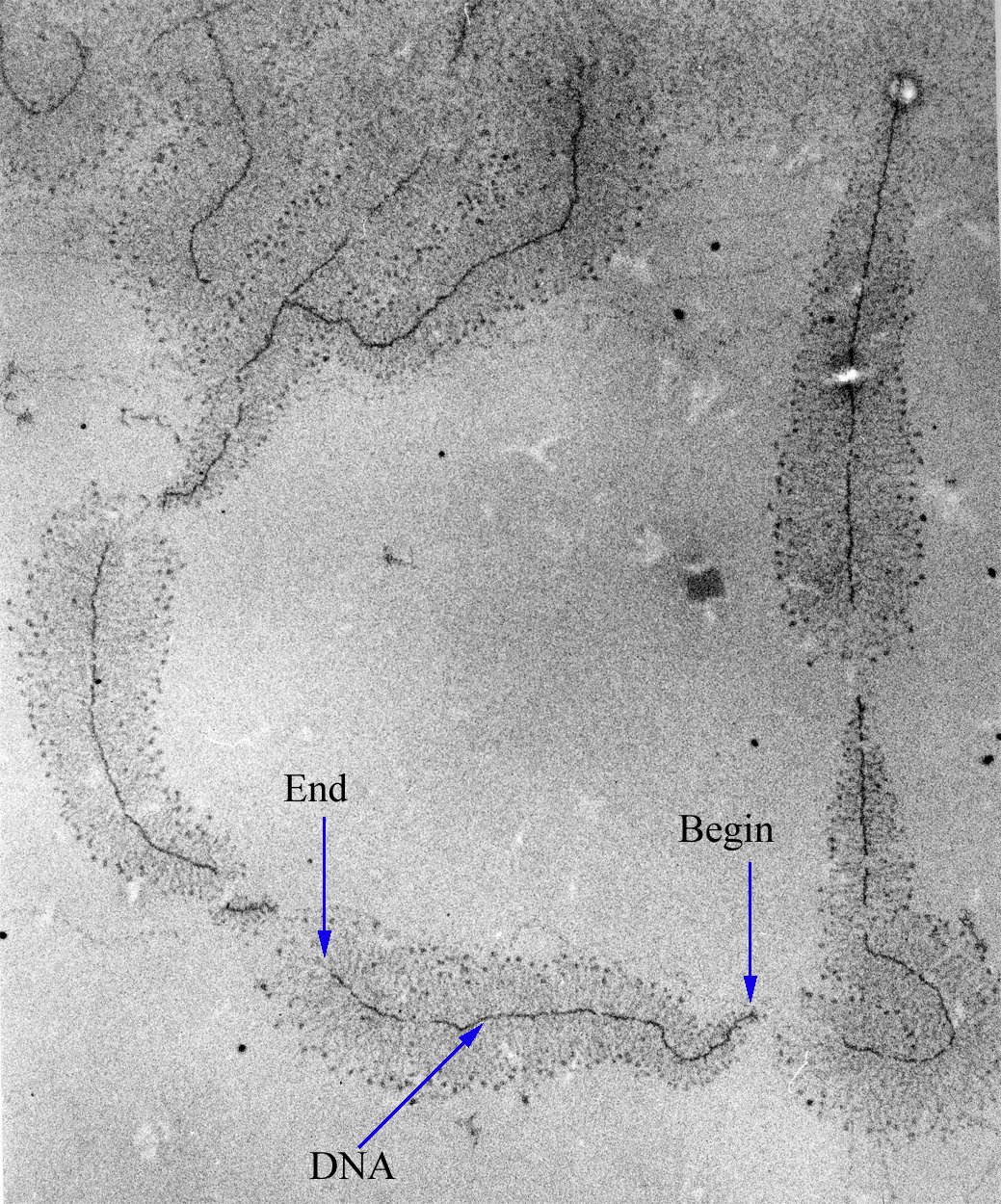
Control of the process of gene transcription affects patterns of gene expression and thereby allows a cell to adapt to a changing environment, perform specialized roles within an organism, and maintain basic metabolic processes necessary for survival. Therefore, it is hardly surprising that the activity of RNAP is both complex and highly regulated. In Escherichia coli bacteria, more than 100 factors have been identified which modify the activity of RNAP.[4]
RNAP can initiate transcription at specific DNA sequences known as promoters. It then produces an RNA chain which is complementary to the template DNA strand. The process of adding nucleotides to the RNA strand is known as elongation; In eukaryotes, RNAP can build chains as long as 2.4 million nucleosides (the full length of the dystrophin gene). RNAP will preferentially release its RNA transcript at specific DNA sequences encoded at the end of genes known as terminators.
Products of RNAP include:
- Messenger RNA (mRNA)—template for the synthesis of proteins by ribosomes.
- Non-coding RNA or "RNA genes"—a broad class of genes that encode RNA that is not translated into protein. The most prominent examples of RNA genes are transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation. However, since the late 1990s, many new RNA genes have been found, and thus RNA genes may play a much more significant role than previously thought.
- Transfer RNA (tRNA)—transfers specific amino acids to growing polypeptide chains at the ribosomal site of protein synthesis during translation
- Ribosomal RNA (rRNA)—a component of ribosomes
- Micro RNA—regulates gene activity
- Catalytic RNA (Ribozyme)—enzymatically active RNA molecules
RNAP accomplishes de novo synthesis. It is able to do this because specific interactions with the initiating nucleotide hold RNAP rigidly in place, facilitating chemical attack on the incoming nucleotide. Such specific interactions explain why RNAP prefers to start transcripts with ATP (followed by GTP, UTP, and then CTP). In contrast to DNA polymerase, RNAP includes helicase activity, therefore no separate enzyme is needed to unwind DNA.
RNA polymerase action
Binding and initiation
RNA Polymerase binding involves the α subunit recognizing the upstream element (-40 to -70 base pairs) in DNA, as well as the σ factor recognizing the -10 to -35 region. There are numerous σ factors that regulate gene expression. For example, σ70 is expressed under normal conditions and allows RNAP binding to house-keeping genes, while σ32 elicits RNAP binding to heat-shock genes.
After binding to the DNA, the RNA polymerase switches from a closed complex to an open complex. This change involves the separation of the DNA strands to form a unwound section of DNA of approximately 13bp. Ribonucleotides are base-paired to the template DNA strand, according to Watson-Crick base-pairing interactions. Supercoiling plays an important part in polymerase activity because of the unwinding and rewinding of DNA. Because regions of DNA in front of RNAP are unwound, there is compensatory positive supercoils. Regions behind RNAP are rewound and negative supercoils are present.
Elongation
Transcription elongation involves the further addition of ribonucleotides and the change of the open complex to the transcriptional complex. RNAP cannot start forming full length transcripts because of its strong binding to promoter. Transcription at this stage primarily results in short RNA fragments of around 9 bp in a process known as abortive transcription. Once the RNAP starts forming longer transcripts it clears the promoter. At this point, the -10 to -35 promoter region is disrupted, and the σ factor falls off RNAP. This allows the rest of the RNAP complex to move forward, as the σ factor held the RNAP complex in place.
The 17 bp transcriptional complex has an 8 bp DNA-RNA hybrid, that is, 8 base-pairs involve the RNA transcript bound to the DNA template strand. As transcription progresses, ribonucleotides are added to the 3' end of the RNA transcript and the RNAP complex moves along the DNA. Although RNAP does not seem to have the 3'exonuclease activity that characterizes the proofreading activity found in DNA polymerase, there is evidence of that RNAP will halt at mismatched base-pairs and correct it.
The addition of ribonucleotides to the RNA transcript has a very similar mechanism to DNA polymerization - it is believed that these polymerases are evolutionarily related. Aspartyl (asp) residues in the RNAP will hold onto Mg2+ ions, which will in turn coordinate the phosphates of the ribonucleotides. The first Mg2+ will hold onto the α-phosphate of the NTP to be added. This allows the nucleophilic attack of the 3'OH from the RNA transcript, adding an additional NTP to the chain. The second Mg2+ will hold onto the pyrophosphate of the NTP. The overall reaction equation is:
(NMP)n + NTP --> (NMP)n+1 + PPi
Termination
Termination of RNA transcription can be rho-independent or rho-dependent:
Rho-independent transcription termination is the termination of transcription without the aid of the rho protein. Transcription of a palindromic region of DNA causes the formation of a hairpin structure from the RNA transcription looping and binding upon itself. This hairpin structure is often rich in G-C base-pairs, making it more stable than the DNA-RNA hybrid itself. As a result, the 8bp DNA-RNA hybrid in the transcription complex shifts to a 4bp hybrid. Coincidentally, these last 4 base-pairs are weak A-U base-pairs, and the entire RNA transcript will fall off.[5]
RNA polymerase in bacteria
In bacteria, the same enzyme catalyzes the synthesis of mRNA and ncRNA.
RNAP is a relatively large molecule. The core enzyme has 5 subunits (~400 kDa):
- α2: the two α subunits assemble the enzyme and recognize regulatory factors. Each subunit has two domains: αCTD (C-Terminal domain) binds the UP element of the extended promoter, and αNTD (N-terminal domain) binds the rest of the polymerase.
- β: this has the polymerase activity (catalyzes the synthesis of RNA) which includes chain initiation and elongation.
- β': binds to DNA (nonspecifically).
- ω: restores denatured RNA polymerase to its functional form in vitro. It has been observed to offer a protective/chaperone function to the β' subunit in Mycobacterium smegmatis. Now known to promote assembly.
In order to bind promoter-specific regions, the core enzyme requires another subunit, sigma (σ). The sigma factor greatly reduces the affinity of RNAP for nonspecific DNA while increasing specificity for certain promoter regions, depending on the sigma factor. That way, transcription is initiated at the right region. The complete holoenzyme therefore has 6 subunits: α2ββ'σω (~480 kDa). The structure of RNAP exhibits a groove with a length of 55 Å (5.5 nm) and a diameter of 25 Å (2.5 nm). This groove fits well the 20 Å (2 nm) double strand of DNA. The 55 Å (5.5 nm) length can accept 16 nucleotides.
When not in use RNA polymerase binds to low affinity sites to allow rapid exchange for an active promoter site when one opens. RNA polymerase holoenzyme, therefore, does not freely float around in the cell when not in use.
Transcriptional cofactors
There are a number of proteins which can bind to RNAP and modify its behavior. For instance, greA and greB from E. coli can enhance the ability of RNAP to cleave the RNA template near the growing end of the chain. This cleavage can rescue a stalled polymerase molecule, and is likely involved in proofreading the occasional mistakes made by RNAP. A separate cofactor, Mfd, is involved in transcription-coupled repair, the process in which RNAP recognizes damaged bases in the DNA template and recruits enzymes to restore the DNA. Other cofactors are known to play regulatory roles, i.e. they help RNAP choose whether or not to express certain genes.
RNA polymerase in eukaryotes

Eukaryotes have several types of RNAP, characterized by the type of RNA they synthesize:
- RNA polymerase I synthesizes a pre-rRNA 45S, which matures into 28S, 18S and 5.8S rRNAs which will form the major RNA sections of the ribosome.[6]
- RNA polymerase II synthesizes precursors of mRNAs and most snRNA and microRNAs.[7] This is the most studied type, and due to the high level of control required over transcription a range of transcription factors are required for its binding to promoters.
- RNA polymerase III synthesizes tRNAs, rRNA 5S and other small RNAs found in the nucleus and cytosol.[8]
There are other RNA polymerase types in mitochondria and chloroplasts.
RNA polymerase in archaea
Archaea have a single RNAP that is closely related to the three main eukaryotic polymerases. Thus, it has been speculated that the archaeal polymerase resembles the ancestor of the specialized eukaryotic polymerases.[9]
RNA polymerase in viruses

Many viruses also encode for RNAP. Perhaps the most widely studied viral RNAP is found in bacteriophage T7. This single-subunit RNAP is related to that found in mitochondria and chloroplasts, and shares considerable homology to DNA polymerase.[10] It is believed that most viral polymerases therefore evolved from DNA polymerase and are not directly related to the multi-subunit polymerases described above.
The viral polymerases are diverse, and include some forms which can use RNA as a template instead of DNA. This occurs in negative strand RNA viruses and dsRNA viruses, both of which exist for a portion of their life cycle as double-stranded RNA. However, some positive strand RNA viruses, such as polio, also contain these RNA dependent RNA polymerases.[11]
Functional domains
C-terminal domain of RNA polymerase
Transcription Initiation
The carboxy-terminal domain (CTD) of RNA polymerase II is that portion of the polymerase which is involved in the initiation of DNA transcription. The CTD typically consists of up to 52 repeats of the sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser [12]. The transcription factor TFIIH is a kinase and will hyperphosphorylate the CTD of RNAP, and in doing so, causes the RNAP complex to move away from the initiation site.
5'Capping
The carboxy-terminal domain is also the binding site of the cap-synthesizing and cap-binding complex. In eukaryotes, after transcription of the 5' end of an RNA transcript, the cap-synthesizing complex on the CTD will remove the gamma-phosphate from the 5'phosphate and attach a GMP, forming a 5',5'-triphosphate linkage. The synthesizing complex falls off and the cap then binds to the cap-binding complex (CBC), which is bound to the CTD.
The 5'cap of eukaryotic RNA transcripts is important for binding of the RNA transcript to the ribosome during translation, to the CTD of RNAP, and prevents RNA degradation.
Spliceosome
The carboxy-terminal domain is also the binding site for spliceosome factors that are part of RNA splicing. These allow for the splicing and removal of introns (in the form of a lariat structure) during RNA transcription.
Mutation in the CTD
Major studies have been carried out in which knockout of particular amino acids was achieved in the CTD. The results indicate that RNA polymerase II CTD truncation mutations affect the ability to induce transcription of a subset of genes in vivo, and the lack of response to induction maps to the upstream activating sequences of these genes.
RNA polymerase purification
RNA polymerase can be isolated in the following ways:
- By a phosphocellulose column.[13]
- By glycerol gradient centrifugation.[14]
- By a DNA column.
- By an Ion exchange column.[15]
And also combinations of the above techniques.
See also
References
- ↑ Jerard Hurwitz (2005). "The Discovery of RNA Polymerase". Journal of Biological Chemistry. 280 (52): 42477–85. doi:10.1074/jbc.X500006200. PMID 16230341. Unknown parameter
|month=ignored (help) - ↑ Nobel Prize 1959
- ↑ Nobel Prize in Chemistry 2006
- ↑ Akira Ishihama (2000). "Functional modulation of Escherichia coli RNA polymerase". 54: 499–518. PMID 11018136.
- ↑ Farnham PJ (1981). "Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro". Nucleic Acids Res. 9 (3): 563–77. PMID 7012794. Unknown parameter
|month=ignored (help); Unknown parameter|coauthors=ignored (help) - ↑ Grummt I. (1999). "Regulation of mammalian ribosomal gene transcription by RNA polymerase I.". Prog Nucleic Acid Res Mol Biol. 62: 109–54. PMID 9932453.
- ↑ Lee Y (2004). "MicroRNA genes are transcribed by RNA polymerase II". EMBO J. 23 (20): 4051–60. PMID 15372072. Unknown parameter
|month=ignored (help); Unknown parameter|coauthors=ignored (help) - ↑ Willis IM. (1993). "RNA polymerase III. Genes, factors and transcriptional specificity". Eur J Biochem. 212 (1): 1–11. PMID 8444147. Unknown parameter
|month=ignored (help) - ↑ D Langer, J Hain, P Thuriaux and W Zillig (1995) Transcription in Archaea: Similarity to that in Eucarya PNAS 92 5768-5772
- ↑ Hedtke et al. (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 227 809-811
- ↑ Paul Ahlquist (2002) RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing. Science 296 1270-1273
- ↑ Anton Meinhart1 (2004). "Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors". Nature. 430 (6996): 223–226. doi:10.1038/nature02679. PMID 15241417. Unknown parameter
|coauthors=ignored (help); Unknown parameter|month=ignored (help) - ↑ Kelly JL (1986). "Yeast mitochondrial RNA polymerase. Purification and properties of the catalytic subunit". J Biol Chem. 261 (22): 10340–7. PMID 3525543. Unknown parameter
|month=ignored (help); Unknown parameter|coauthors=ignored (help) - ↑ Honda A; et al. (1990). "Purification and molecular structure of RNA polymerase from influenza virus A/PR8". J Biochem (Tokyo). 107 (4): 624–8. PMID 2358436. Unknown parameter
|month=ignored (help) - ↑ Hager et al. (1990) Use of Mono Q High-Resolution Ion-Exchange Chromatography To Obtain Highly Pure and Active Escherichia coli RNA Polymerase Biochemistry 29 7890-7894
External links
- DNAi - DNA Interactive, including information and Flash clips on RNA Polymerase.
- RNA+Polymerase at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 2.7.7.6
Template:Polymerases Template:SIB da:RNA polymerase de:RNA-Polymerase el:RNA πολυμεράση it:RNA polimerasi (DNA-dipendente) he:RNA פולימראז nl:RNA-polymerase oc:ARN polimerasa fi:RNA-polymeraasi uk:РНК-полімераза