West nile virus
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
WNV is an enveloped positive-sense ssRNA virus of 11000 base pairs (bp) that is considered a member of the Japanese encephalitis serocomplex. It belongs to the genus Flavivirus and family Flaviviridae. Its RNA encodes structural and non-structural proteins. Although 7 lineages of WNV have been described, only lineage 1 and 2 are clinically significant. The viral natural reservoir includes many species, such as humans, horses, dogs, and cats; but the main natural reservoir is birds.
Taxonomy
Viruses; ssRNA viruses; ssRNA positive-strand viruses, no DNA stage; Flaviviridae; Flavivirus; Japanese encephalitis virus group[1]
Biology
 |
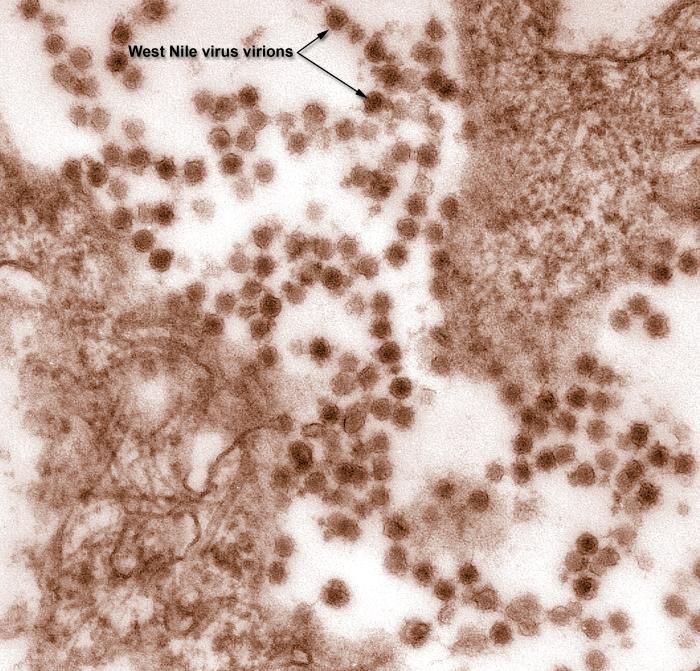 |
WNV is a member of Japanese encephalitis serocomplex and belongs to the genus Flavivirus, family Flaviviridae. Other species of the this serocomplex include the St Louis encephalitis virus and the Japanese encephalitis virus.[4]
The WNV has an icosahedral symmetry, with a smooth surface.[5] It is an enveloped virus with a nucleocapsid core built of RNA and capsid proteins. Its genome is contained in a single-stranded RNA of about 11000 bp.[6] It contains a single open reading frame (ORF), a 5' untranslated region (UTR), and another 3' region which is also not translated. The ORF contains a single polyprotein that produces 3 smaller types of structure proteins and 7 of non-structural proteins following processing and translation.
- Structural proteins are responsible for the formation of the viral particle and include:
- Envelope proteins
- Membrane proteins
- C proteins
- Non-structural proteins are responsible for viral replication, evasion of the immune system, and assembly of virions. They include:
- NS1
- NS2A
- NS2B
- NS3
- NS4A
- NS4B
- NS5
The WNV may be classified in 7 phylogenetic lineages. Of these, only 1 and 2 have been identified as causative agents of disease in humans and are considered clinically significant.[7][8][9][10][11][12][13][14]
- Lineage 1: Widespread, isolates from Europe, America, Middle East, India, Africa, and Australia
- Lingeage 2: Southern Africa, Madagascar, and Europe
Natural reservoir
Although WNV can infect humans and numerous animals, birds are its main natural reservoir.[4][6]
References
- ↑ "West Nile Virus".
- ↑ "http://phil.cdc.gov/phil/details.asp". External link in
|title=(help) - ↑ "http://phil.cdc.gov/phil/details.asp". External link in
|title=(help) - ↑ 4.0 4.1 Petersen LR, Brault AC, Nasci RS (2013). "West Nile virus: review of the literature". JAMA. 310 (3): 308–15. doi:10.1001/jama.2013.8042. PMID 23860989.
- ↑ Mukhopadhyay, S. (2003). "Structure of West Nile Virus". Science. 302 (5643): 248–248. doi:10.1126/science.1089316. ISSN 0036-8075.
- ↑ 6.0 6.1 Campbell, Grant L; Marfin, Anthony A; Lanciotti, Robert S; Gubler, Duane J (2002). "West Nile virus". The Lancet Infectious Diseases. 2 (9): 519–529. doi:10.1016/S1473-3099(02)00368-7. ISSN 1473-3099.
- ↑ "West Nile Virus" (PDF).
- ↑ Miller DL, Mauel MJ, Baldwin C, Burtle G, Ingram D, Hines ME; et al. (2003). "West Nile virus in farmed alligators". Emerg Infect Dis. 9 (7): 794–9. doi:10.3201/eid0907.030085. PMC 3023431. PMID 12890319.
- ↑ Bakonyi T, Ivanics E, Erdélyi K, Ursu K, Ferenczi E, Weissenböck H; et al. (2006). "Lineage 1 and 2 strains of encephalitic West Nile virus, central Europe". Emerg Infect Dis. 12 (4): 618–23. doi:10.3201/eid1204.051379. PMC 3294705. PMID 16704810.
- ↑ Charrel RN, Brault AC, Gallian P, Lemasson JJ, Murgue B, Murri S; et al. (2003). "Evolutionary relationship between Old World West Nile virus strains. Evidence for viral gene flow between Africa, the Middle East, and Europe". Virology. 315 (2): 381–8. PMID 14585341.
- ↑ Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R; et al. (2002). "Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East". Virology. 298 (1): 96–105. PMID 12093177.
- ↑ Papa A, Xanthopoulou K, Gewehr S, Mourelatos S (2011). "Detection of West Nile virus lineage 2 in mosquitoes during a human outbreak in Greece". Clin Microbiol Infect. 17 (8): 1176–80. doi:10.1111/j.1469-0691.2010.03438.x. PMID 21781205.
- ↑ Savini G, Capelli G, Monaco F, Polci A, Russo F, Di Gennaro A; et al. (2012). "Evidence of West Nile virus lineage 2 circulation in Northern Italy". Vet Microbiol. 158 (3–4): 267–73. doi:10.1016/j.vetmic.2012.02.018. PMID 22406344.
- ↑ Valiakos G, Touloudi A, Iacovakis C, Athanasiou L, Birtsas P, Spyrou V; et al. (2011). "Molecular detection and phylogenetic analysis of West Nile virus lineage 2 in sedentary wild birds (Eurasian magpie), Greece, 2010". Euro Surveill. 16 (18). PMID 21586266.