Mothers against decapentaplegic homolog 4: Difference between revisions
m (Reverted edits by 187.140.208.72 (talk) (HG) (3.3.3)) |
(Reverted 1 edit by 128.250.0.107 (talk): Do not spam your own research all over. (TW)) |
||
| Line 1: | Line 1: | ||
{{Infobox_gene}} | {{Infobox_gene}} | ||
''' | '''SMAD4''', also called '''SMAD family member 4''', '''Mothers against decapentaplegic homolog 4''', or '''DPC4 (Deleted in Pancreatic Cancer-4)''' is a highly-conserved protein present in all [[metazoans]]. It belongs to the [[SMAD (protein)|SMAD]] family of [[transcription factor]] proteins, which act as mediators of TGF-β signal transduction. The [[transforming growth factor beta|TGFβ]] family of cytokines regulates critical processes during the lifecycle of metazoans, with important roles during embryo development, tissue homeostasis, regeneration, and immune regulation.<ref>{{Cite journal|last=Massagué|first=Joan|title=TGFβ signalling in context|url=http://www.nature.com/doifinder/10.1038/nrm3434|journal=Nature Reviews Molecular Cell Biology|language=En|volume=13|issue=10|pages=616–630|doi=10.1038/nrm3434|issn=1471-0080|year=2012|pmc=4027049}}</ref> | ||
SMAD 4 belongs to the [[co-SMAD]] group, the second class of the SMAD family. SMAD4 is the only known co-SMAD in most metazoans. It also belongs to the [[Darwin family]] of proteins that modulate members of the [[transforming growth factor beta|TGFβ]] protein superfamily, a family of proteins that all play a role in the regulation of cellular responses. Mammalian SMAD4 is a [[Homology (biology)|homolog]] of the ''[[Drosophila]]'' protein "[[Mothers against decapentaplegic]]" named Medea.<ref name="Massagué1998">{{cite journal|last1=Massagué|first1=J.|title=TGF-β SIGNAL TRANSDUCTION|journal=Annual Review of Biochemistry|volume=67|issue=1|year=1998|pages=753–791|issn=0066-4154|doi=10.1146/annurev.biochem.67.1.753}}</ref> | |||
SMAD 4 belongs to the [[co-SMAD]] group, the second class of the SMAD family. | |||
= | SMAD4 interacts with R-Smads, such as [[SMAD2]], [[SMAD3]], [[SMAD1]], [[SMAD5]] and [[SMAD8]] (also called SMAD9) to form heterotrimeric complexes. Once in the nucleus, the complex of SMAD4 and two R-SMADS binds to [[DNA]] and regulates the expression of different genes depending on the cellular context.<ref name="Massagué1998"/> Intracellular reactions involving SMAD4 are triggered by the binding, on the surface of the cells, of growth factors from the [[TGFβ]] family. The sequence of intracellular reactions involving SMADS is called the SMAD pathway or the transforming growth factor beta (TGF-β) pathway since the sequence starts with the recognition of TGF-β by cells. | ||
== Gene == | |||
In mammals, SMAD4 is coded by a gene located on [[chromosome 18]]. In humans, the ''SMAD4'' gene contains 54 829 base pairs and is located from pair n° 51,030,212 to pair 51,085,041 in the region 21.1 of the chromosome 18.<ref name="entrez">{{cite web | work = Entrez Gene | title = SMAD4 SMAD family member 4 | url = https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4089 | accessdate = }}</ref><ref name="gene library">{{cite web |title= SMAD 4 | work = The Genetics Home Reference Website | url = http://ghr.nlm.nih.gov/gene/SMAD4 }}</ref> | |||
[[File:Chromosome 18.svg|Chromosome 18|File:Chromosome 18.svg|thumb|Chromosome 18|center|Pattern of the chromosome 18 in ''Homo sapiens''. The ''SMAD 4'' gene is located on the long arm of the chromosome, at locus 21.1. This locus corresponds to the black stripe between the regions 12.3 and 21.2.]] | |||
[[ | == Protein == | ||
SMAD4 is a 552 amino-acid [[polypeptide]] with a molecular weight of 60.439 [[Unified atomic mass unit|Da]]. SMAD4 has two functional domains known as [[MH1 domain|MH1]] and [[MH2 domain|MH2]]. | |||
SMAD 4 | [[File:PDB 1dd1 EBI.jpg|PDB 1dd1 EBI|File:PDB 1dd1 EBI.jpg|thumb|PDB 1dd1 EBI|SMAD 4 is composed of three major domains, including MH1 (up), MH2 (down) and a linking domain (right).]]The complex of two SMAD3 (or of two SMAD2) and one SMAD4 binds directly to DNA though interactions of their MH1 domains. These complexes are recruited to sites throughout the genome by cell lineage-defining transcription factors (LDTFs) that determine the context-dependent nature of TGF-β action. Early insights into the DNA binding specificity of Smad proteins came from oligonucleotide binding screens, which identified the palindromic duplex 5’–GTCTAGAC–3’ as a high affinity binding sequence for SMAD3 and SMAD4 MH1 domains.<ref>{{cite journal|date=Mar 1998|title=Human Smad3 and Smad4 are sequence-specific transcription activators|journal=Molecular Cell|volume=1|issue=4|pages=611–617|doi=10.1016/s1097-2765(00)80061-1|pmid=9660945|vauthors=Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE}}</ref> Other motifs have also been identified in promoters and enhancers. These additional sites contain the CAGCC motif and the GGC(GC)|(CG) consensus sequences, the latter also known as 5GC sites.<ref name="Martin2017" >{{Cite journal|last=Martin-Malpartida|first=Pau|last2=Batet|first2=Marta|last3=Kaczmarska|first3=Zuzanna|last4=Freier|first4=Regina|last5=Gomes|first5=Tiago|last6=Aragón|first6=Eric|last7=Zou|first7=Yilong|last8=Wang|first8=Qiong|last9=Xi|first9=Qiaoran|date=2017-12-12|title=Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors|url=http://www.nature.com/articles/s41467-017-02054-6|journal=Nature Communications|language=En|volume=8|issue=1|doi=10.1038/s41467-017-02054-6|issn=2041-1723}}</ref> The 5GC-motifs are highly represented as clusters of sites, in SMAD-bound regions genome-wide. These clusters can also contain CAG(AC)|(CC) sites. SMAD3/SMAD4 complex also binds to the TPA-responsive gene promoter elements, which have the sequence motif TGAGTCAG.<ref name="ZhangFeng1998">{{cite journal|last1=Zhang|first1=Ying|last2=Feng|first2=Xin-Hua|last3=Derynck|first3=Rik|title=Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription|journal=Nature|volume=394|issue=6696|year=1998|pages=909–913|issn=0028-0836|doi=10.1038/29814}}</ref> | ||
== Structures == | |||
===MH1 domain complexes with DNA motifs=== | |||
The | The first structure of SMAD4 bound to DNA was the complex with the palindromic GTCTAGAC motif.<ref name="BaburajendranJauch2011">{{cite journal|last1=Baburajendran|first1=Nithya|last2=Jauch|first2=Ralf|last3=Tan|first3=Clara Yueh Zhen|last4=Narasimhan|first4=Kamesh|last5=Kolatkar|first5=Prasanna R.|title=Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers|journal=Nucleic Acids Research|volume=39|issue=18|year=2011|pages=8213–8222|issn=1362-4962|doi=10.1093/nar/gkr500}}</ref> Recently, the structures of SMAD4 MH1 domain bound to several 5GC motifs have also been determined. In all complexes, the interaction with the DNA involves a conserved β-hairpin present in the MH1 domain. The hairpin is partially flexible in solution and its high degree of conformational flexibility allows recognition of the different 5-bp sequences. Efficient interactions with GC-sites occur only if a G nucleotide is located deep in the major grove, and establishes hydrogen bonds with the guanidinium group of Arg81. This interaction facilitates a complementary surface contact between the Smad DNA-binding hairpin and the major groove of the DNA. Other direct interactions involve Lys88 and Gln83. The X-ray crystal structure of the ''[[Trichoplax adhaerens]]'' SMAD4 MH1 domains bound to the GGCGC motif indicates a high conservation of this interaction in metazoans.<ref name="Martin2017"/> | ||
[[File:Chimera render of Smad4 protein from PDB 5MEZ.png|thumb|Smad4 MH1 domain bound to the GGCT DNA motif, from {{PDB|5MEZ}}]] | |||
[[File:Smad4 bound to the GGCGC motif (close-up).png|thumb|Close-up view of the Smad4 MH1 domain bound to the GGCGC DNA motif, from {{PDB|5MEY}}]] | |||
[[File:Smad4 bound to the GGCGC motif.png|thumb|Smad4 MH1 domain bound to the GGCGC DNA motif, from {{PDB|5MEY}}]] | |||
The MH2 domain, corresponding to the [[C-terminus]], is responsible for receptor recognition | ===MH2 domain complexes=== | ||
The MH2 domain, corresponding to the [[C-terminus]], is responsible for receptor recognition and association with other SMADs. It interacts with the R-SMADS MH2 domain and forms [[heterodimers]] and [[heterotrimers]]. Some tumor mutations detected in SMAD4 enhance interactions between the MH1 and MH2 domains.<ref name="HataLo1997">{{cite journal|last1=Hata|first1=Akiko|last2=Lo|first2=Roger S.|last3=Wotton|first3=David|last4=Lagna|first4=Giorgio|last5=Massagué|first5=Joan|title=Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4|journal=Nature|volume=388|issue=6637|year=1997|pages=82–87|issn=0028-0836|doi=10.1038/40424}}</ref> | |||
== Nomenclature and origin of name == | == Nomenclature and origin of name == | ||
SMADs are highly conserved across species, especially in the [[N terminus|N terminal]] [[MH1 domain]] and the [[C terminus|C terminal]] [[MH2 domain]]. | SMADs are highly conserved across species, especially in the [[N terminus|N terminal]] [[MH1 domain]] and the [[C terminus|C terminal]] [[MH2 domain]]. | ||
The SMAD proteins are homologs of both the ''Drosophila'' protein MAD and the ''[[Caenorhabditis elegans|C. elegans]]'' protein SMA. The name is a combination of the two. During ''Drosophila'' research, it was found that a mutation in the gene <!--Do not enclose a qualifier - or separate a predicate from its subject - with commas.-->''MAD'' in the mother repressed the gene ''[[decapentaplegic]]'' in the embryo. The phrase "Mothers against" was added, since mothers often form organizations opposing various issues, e.g. [[Mothers Against Drunk Driving]] (MADD), reflecting "the maternal-effect enhancement of [[decapentaplegic|dpp]]";<ref name="pmid7768443">{{cite journal | vauthors = Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM | title = Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster | journal = Genetics | volume = 139 | issue = 3 | pages = 1347–58 | date = Mar 1995 | pmid = 7768443 | pmc = 1206461 | doi = | url = http://www.genetics.org/content/139/3/1347.full.pdf+html }}</ref> and based on a tradition of unusual naming within the research community.<ref>{{cite web | url = https://psmag.com/sonic-hedgehog-dicer-and-the-problem-with-naming-genes-113c58df8f7a#.os08udsyk | title = Sonic Hedgehog, DICER, and the Problem With Naming Genes | date= 26 September 2014 | first = Michael | last = White | name-list-format = vanc | work = Pacific Standard }}</ref> | The SMAD proteins are homologs of both the ''Drosophila'' protein MAD and the ''[[Caenorhabditis elegans|C. elegans]]'' protein SMA. The name is a combination of the two. During ''Drosophila'' research, it was found that a mutation in the gene <!--Do not enclose a qualifier - or separate a predicate from its subject - with commas.-->''MAD'' in the mother repressed the gene ''[[decapentaplegic]]'' in the embryo. The phrase "Mothers against" was added, since mothers often form organizations opposing various issues, e.g. [[Mothers Against Drunk Driving]] (MADD), reflecting "the maternal-effect enhancement of [[decapentaplegic|dpp]]";<ref name="pmid7768443">{{cite journal | vauthors = Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM | title = Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster | journal = Genetics | volume = 139 | issue = 3 | pages = 1347–58 | date = Mar 1995 | pmid = 7768443 | pmc = 1206461 | doi = | url = http://www.genetics.org/content/139/3/1347.full.pdf+html }}</ref> and based on a tradition of unusual naming within the research community.<ref>{{cite web | url = https://psmag.com/sonic-hedgehog-dicer-and-the-problem-with-naming-genes-113c58df8f7a#.os08udsyk | title = Sonic Hedgehog, DICER, and the Problem With Naming Genes | date= 26 September 2014 | first = Michael | last = White | name-list-format = vanc | work = Pacific Standard }}</ref> SMAD4 is also known as DPC4, JIP or MADH4. | ||
==Function and action mechanism== | ==Function and action mechanism== | ||
SMAD4 is a protein defined as an essential effector in the SMAD pathway. SMAD4 serves as a mediator between extracellular growth factors from the TGFβ family and genes inside the cell [[Cell nucleus|nucleus]]. The abbreviation ''co'' in co-SMAD stands for ''common mediator''. SMAD4 is also defined as a signal transducer. | |||
In the TGF-β pathway, TGF-β dimers are recognized by a transmembrane receptor, known as type II receptor. Once the type II receptor is activated by the binding of TGF-β, it phosphorylates a type I receptor. Type I receptor is also a [[cell surface receptor]]. This receptor then phosphorylates intracellular receptor regulated SMADS (R-SMADS) such as | In the TGF-β pathway, TGF-β dimers are recognized by a transmembrane receptor, known as type II receptor. Once the type II receptor is activated by the binding of TGF-β, it phosphorylates a type I receptor. Type I receptor is also a [[cell surface receptor]]. This receptor then phosphorylates intracellular receptor regulated SMADS (R-SMADS) such as SMAD2 or SMAD3. The phosphorylated R-SMADS then bind to SMAD4. The R-SMADs-SMAD4 association is a [[heteromeric]] complex. This complex is going to move from the cytoplasm to the nucleus: it is the translocation. SMAD4 may form heterotrimeric, heterohexameric or heterodimeric complexes with R-SMADS. | ||
SMAD4 is a substrate of the [[Extracellular signal-regulated kinases|Erk]]/[[MAPK]] kinase <ref>{{cite journal | vauthors = Roelen BA, Cohen OS, Raychowdhury MK, Chadee DN, Zhang Y, Kyriakis JM, Alessandrini AA, Lin HY | title = Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation | journal = American Journal of Physiology. Cell Physiology | volume = 285 | issue = 4 | date = Oct 2003 | pmid = 12801888 | doi = 10.1152/ajpcell.00053.2003 | pages=C823–30}}</ref> and [[GSK3]].<ref>{{cite journal | vauthors = Demagny H, Araki T, De Robertis EM | title = The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-β signaling | journal = Cell Reports | volume = 9 | issue = 2 | date = Oct 2014 | pmid = 25373906 | doi = 10.1016/j.celrep.2014.09.020 | pages=688–700}}</ref> The FGF ([[Fibroblast Growth Factor]]) pathway stimulation leads to Smad4 [[phosphorylation]] by [[Extracellular signal-regulated kinases|Erk]] of the canonical [[MAPK]] site located at Threonine 277. This phosphorylation event has a dual effect on Smad4 activity. First, it allows Smad4 to reach its peak of transcriptional activity by activating a [[growth factor]]-regulated transcription activation domain located in the Smad4 linker region, SAD (Smad-Activation Domain).<ref>{{cite journal|last1=de Caestecker|first1=Mark P.|title=The Smad4 Activation Domain (SAD) Is a Proline-rich, p300-dependent Transcriptional Activation Domain|journal=The Journal of Biological Chemistry|volume=275|issue=3|pages=2115–2122|doi=10.1074/jbc.275.3.2115|pmid=10636916|year=2000}}</ref> Second, [[MAPK]] primes Smad4 for [[GSK3]]-mediated phosphorylations that cause transcriptional inhibition and also generate a phosphodegron used as a docking site by the ubiquitin [[E3 ligase]] Beta-transducin Repeat Containing ([[BTRC (gene)|beta-TrCP]]) that polyubiquitinates Smad4 and targets it for degradation in the [[proteasome]].<ref>{{cite journal | last1 =Demagny | first1 = Hadrien | last2 = De Robertis | first2 = Edward M.|title=Smad4/DPC4: a Barrier against Tumor Progression driven by RTK/Ras/Erk and Wnt/GSK3 signaling | journal = Molecular & Cellular Oncology | volume = 3 | issue = 2 | doi = 10.4161/23723556.2014.989133 | name-list-format = vanc | pages=e989133| year = 2015 | pmc = 4905428 }}</ref> Smad4 [[GSK3]] phosphorylations have been proposed to regulate the protein stability during pancreatic and [[colon cancer]] progression.<ref>{{cite journal | last1 = Demagny | first1 = Hadrien | last2 = De Robertis | first2 = Edward M. | title = Point Mutations in the Tumor Suppressor Smad4/DPC4 Enhance its Phosphorylation by GSK3 and Reversibly Inactivate TGF-β Signaling | journal = Molecular & Cellular Oncology | doi = 10.1080/23723556.2015.1025181 | name-list-format = vanc | volume=3 | pages=e1025181| year = 2015 | pmc = 4845174 }}</ref> | |||
In the nucleus the heteromeric complex binds promoters and interact with transcriptional activators. [[SMAD3]]/SMAD4 complexes can directly bind the SBE. These associations are weak and require additional [[transcription factor]]s such as members of the [[AP-1 (transcription factor)|AP-1]] family, [[TFE3]] and [[FoxG1]] to regulate [[gene expression]].<ref>{{cite journal | vauthors = Inman GJ | title = Linking Smads and transcriptional activation | journal = The Biochemical Journal | volume = 386 | issue = Pt 1 | pages = e1–e3 | date = Feb 2005 | pmid = 15702493 | pmc=1134782 | doi=10.1042/bj20042133}}</ref> | In the nucleus the heteromeric complex binds promoters and interact with transcriptional activators. [[SMAD3]]/SMAD4 complexes can directly bind the SBE. These associations are weak and require additional [[transcription factor]]s such as members of the [[AP-1 (transcription factor)|AP-1]] family, [[TFE3]] and [[FoxG1]] to regulate [[gene expression]].<ref>{{cite journal | vauthors = Inman GJ | title = Linking Smads and transcriptional activation | journal = The Biochemical Journal | volume = 386 | issue = Pt 1 | pages = e1–e3 | date = Feb 2005 | pmid = 15702493 | pmc=1134782 | doi=10.1042/bj20042133}}</ref> | ||
| Line 50: | Line 55: | ||
SMAD4, is often found mutated in many cancers. The mutation can be inherited or acquired during an individual’s lifetime. | SMAD4, is often found mutated in many cancers. The mutation can be inherited or acquired during an individual’s lifetime. | ||
If inherited, the mutation affects both [[somatic (biology)|somatic]] and | If inherited, the mutation affects both [[somatic (biology)|somatic]] cells and cells of the reproductive organs. If the ''SMAD 4'' mutation is acquired, it will only exist in certain somatic cells. Indeed, SMAD 4 is not synthesized by all cells. The protein is present in skin, pancreatic, colon, uterus and epithelial cells. It is also produced by [[fibroblasts]]. | ||
The functional SMAD 4 participates in the regulation of the TGF-β signal transduction pathway, which negatively regulates growth of epithelial cells and the [[extracellular matrix]] (ECM). When the structure of SMAD 4 is altered, expression of the genes involved in cell growth is no longer regulated and cell proliferation can go on without any inhibition. The important number of cell divisions leads to the forming of tumors and then to multiploid [[colorectal cancer]] and [[pancreatic carcinoma]]. It is found inactivated in at least 50% of pancreatic cancers.<ref name="isbn0-7216-0187-1">{{cite book | last1 = Cotran | first1 = Ramzi S. | last2 = Kumar | first2 = Vinay | last3 = Fausto | first3 = Nelson | last4 = Robbins | first4 = Stanley L. | last5 = Abbas | first5 = Abul K. | title = Robbins and Cotran pathologic basis of disease | edition = 7th | publisher = Elsevier Saunders | location = St. Louis, Mo | year = 2005 | pages = | quote = | isbn = 0-7216-0187-1 | name-list-format = vanc }}</ref> | The functional SMAD 4 participates in the regulation of the TGF-β signal transduction pathway, which negatively regulates growth of epithelial cells and the [[extracellular matrix]] (ECM). When the structure of SMAD 4 is altered, expression of the genes involved in cell growth is no longer regulated and cell proliferation can go on without any inhibition. The important number of cell divisions leads to the forming of tumors and then to multiploid [[colorectal cancer]] and [[pancreatic carcinoma]]. It is found inactivated in at least 50% of pancreatic cancers.<ref name="isbn0-7216-0187-1">{{cite book | last1 = Cotran | first1 = Ramzi S. | last2 = Kumar | first2 = Vinay | last3 = Fausto | first3 = Nelson | last4 = Robbins | first4 = Stanley L. | last5 = Abbas | first5 = Abul K. | title = Robbins and Cotran pathologic basis of disease | edition = 7th | publisher = Elsevier Saunders | location = St. Louis, Mo | year = 2005 | pages = | quote = | isbn = 0-7216-0187-1 | name-list-format = vanc }}</ref> | ||
Revision as of 13:52, 31 August 2018
| VALUE_ERROR (nil) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Aliases | |||||||
| External IDs | GeneCards: [1] | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
|
| |||||
| Ensembl |
|
| |||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | n/a | n/a | |||||
| Wikidata | |||||||
| |||||||
SMAD4, also called SMAD family member 4, Mothers against decapentaplegic homolog 4, or DPC4 (Deleted in Pancreatic Cancer-4) is a highly-conserved protein present in all metazoans. It belongs to the SMAD family of transcription factor proteins, which act as mediators of TGF-β signal transduction. The TGFβ family of cytokines regulates critical processes during the lifecycle of metazoans, with important roles during embryo development, tissue homeostasis, regeneration, and immune regulation.[1]
SMAD 4 belongs to the co-SMAD group, the second class of the SMAD family. SMAD4 is the only known co-SMAD in most metazoans. It also belongs to the Darwin family of proteins that modulate members of the TGFβ protein superfamily, a family of proteins that all play a role in the regulation of cellular responses. Mammalian SMAD4 is a homolog of the Drosophila protein "Mothers against decapentaplegic" named Medea.[2]
SMAD4 interacts with R-Smads, such as SMAD2, SMAD3, SMAD1, SMAD5 and SMAD8 (also called SMAD9) to form heterotrimeric complexes. Once in the nucleus, the complex of SMAD4 and two R-SMADS binds to DNA and regulates the expression of different genes depending on the cellular context.[2] Intracellular reactions involving SMAD4 are triggered by the binding, on the surface of the cells, of growth factors from the TGFβ family. The sequence of intracellular reactions involving SMADS is called the SMAD pathway or the transforming growth factor beta (TGF-β) pathway since the sequence starts with the recognition of TGF-β by cells.
Gene
In mammals, SMAD4 is coded by a gene located on chromosome 18. In humans, the SMAD4 gene contains 54 829 base pairs and is located from pair n° 51,030,212 to pair 51,085,041 in the region 21.1 of the chromosome 18.[3][4]
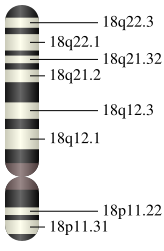
Protein
SMAD4 is a 552 amino-acid polypeptide with a molecular weight of 60.439 Da. SMAD4 has two functional domains known as MH1 and MH2.
The complex of two SMAD3 (or of two SMAD2) and one SMAD4 binds directly to DNA though interactions of their MH1 domains. These complexes are recruited to sites throughout the genome by cell lineage-defining transcription factors (LDTFs) that determine the context-dependent nature of TGF-β action. Early insights into the DNA binding specificity of Smad proteins came from oligonucleotide binding screens, which identified the palindromic duplex 5’–GTCTAGAC–3’ as a high affinity binding sequence for SMAD3 and SMAD4 MH1 domains.[5] Other motifs have also been identified in promoters and enhancers. These additional sites contain the CAGCC motif and the GGC(GC)|(CG) consensus sequences, the latter also known as 5GC sites.[6] The 5GC-motifs are highly represented as clusters of sites, in SMAD-bound regions genome-wide. These clusters can also contain CAG(AC)|(CC) sites. SMAD3/SMAD4 complex also binds to the TPA-responsive gene promoter elements, which have the sequence motif TGAGTCAG.[7]
Structures
MH1 domain complexes with DNA motifs
The first structure of SMAD4 bound to DNA was the complex with the palindromic GTCTAGAC motif.[8] Recently, the structures of SMAD4 MH1 domain bound to several 5GC motifs have also been determined. In all complexes, the interaction with the DNA involves a conserved β-hairpin present in the MH1 domain. The hairpin is partially flexible in solution and its high degree of conformational flexibility allows recognition of the different 5-bp sequences. Efficient interactions with GC-sites occur only if a G nucleotide is located deep in the major grove, and establishes hydrogen bonds with the guanidinium group of Arg81. This interaction facilitates a complementary surface contact between the Smad DNA-binding hairpin and the major groove of the DNA. Other direct interactions involve Lys88 and Gln83. The X-ray crystal structure of the Trichoplax adhaerens SMAD4 MH1 domains bound to the GGCGC motif indicates a high conservation of this interaction in metazoans.[6]
MH2 domain complexes
The MH2 domain, corresponding to the C-terminus, is responsible for receptor recognition and association with other SMADs. It interacts with the R-SMADS MH2 domain and forms heterodimers and heterotrimers. Some tumor mutations detected in SMAD4 enhance interactions between the MH1 and MH2 domains.[9]
Nomenclature and origin of name
SMADs are highly conserved across species, especially in the N terminal MH1 domain and the C terminal MH2 domain. The SMAD proteins are homologs of both the Drosophila protein MAD and the C. elegans protein SMA. The name is a combination of the two. During Drosophila research, it was found that a mutation in the gene MAD in the mother repressed the gene decapentaplegic in the embryo. The phrase "Mothers against" was added, since mothers often form organizations opposing various issues, e.g. Mothers Against Drunk Driving (MADD), reflecting "the maternal-effect enhancement of dpp";[10] and based on a tradition of unusual naming within the research community.[11] SMAD4 is also known as DPC4, JIP or MADH4.
Function and action mechanism
SMAD4 is a protein defined as an essential effector in the SMAD pathway. SMAD4 serves as a mediator between extracellular growth factors from the TGFβ family and genes inside the cell nucleus. The abbreviation co in co-SMAD stands for common mediator. SMAD4 is also defined as a signal transducer.
In the TGF-β pathway, TGF-β dimers are recognized by a transmembrane receptor, known as type II receptor. Once the type II receptor is activated by the binding of TGF-β, it phosphorylates a type I receptor. Type I receptor is also a cell surface receptor. This receptor then phosphorylates intracellular receptor regulated SMADS (R-SMADS) such as SMAD2 or SMAD3. The phosphorylated R-SMADS then bind to SMAD4. The R-SMADs-SMAD4 association is a heteromeric complex. This complex is going to move from the cytoplasm to the nucleus: it is the translocation. SMAD4 may form heterotrimeric, heterohexameric or heterodimeric complexes with R-SMADS.
SMAD4 is a substrate of the Erk/MAPK kinase [12] and GSK3.[13] The FGF (Fibroblast Growth Factor) pathway stimulation leads to Smad4 phosphorylation by Erk of the canonical MAPK site located at Threonine 277. This phosphorylation event has a dual effect on Smad4 activity. First, it allows Smad4 to reach its peak of transcriptional activity by activating a growth factor-regulated transcription activation domain located in the Smad4 linker region, SAD (Smad-Activation Domain).[14] Second, MAPK primes Smad4 for GSK3-mediated phosphorylations that cause transcriptional inhibition and also generate a phosphodegron used as a docking site by the ubiquitin E3 ligase Beta-transducin Repeat Containing (beta-TrCP) that polyubiquitinates Smad4 and targets it for degradation in the proteasome.[15] Smad4 GSK3 phosphorylations have been proposed to regulate the protein stability during pancreatic and colon cancer progression.[16]
In the nucleus the heteromeric complex binds promoters and interact with transcriptional activators. SMAD3/SMAD4 complexes can directly bind the SBE. These associations are weak and require additional transcription factors such as members of the AP-1 family, TFE3 and FoxG1 to regulate gene expression.[17]
Many TGFβ ligands use this pathway and subsequently SMAD4 is involved in many cell functions such as differentiation, apoptosis, gastrulation, embryonic development and the cell cycle.
Clinical significance
Genetic experiments such as gene knockout (KO), which consist in modifying or inactivating a gene, can be carried out in order to see the effects of a dysfunctional SMAD 4 on the study organism. Experiments are often conducted in the house mouse (Mus musculus).
It has been shown that, in mouse KO of SMAD4, the granulosa cells, which secrete hormones and growth factors during the oocyte development, undergo premature luteinization and express lower levels of follicle-stimulating hormone receptors (FSHR) and higher levels of luteinizing hormone receptors (LHR). This may be due in part to impairment of bone morphogenetic protein-7 effects as BMP-7 uses the SMAD4 signaling pathway.[18][19]
Deletions in the genes coding for SMAD1 and SMAD5 have also been linked to metastasic granulosa cell tumors in mice.[20]
SMAD4, is often found mutated in many cancers. The mutation can be inherited or acquired during an individual’s lifetime. If inherited, the mutation affects both somatic cells and cells of the reproductive organs. If the SMAD 4 mutation is acquired, it will only exist in certain somatic cells. Indeed, SMAD 4 is not synthesized by all cells. The protein is present in skin, pancreatic, colon, uterus and epithelial cells. It is also produced by fibroblasts. The functional SMAD 4 participates in the regulation of the TGF-β signal transduction pathway, which negatively regulates growth of epithelial cells and the extracellular matrix (ECM). When the structure of SMAD 4 is altered, expression of the genes involved in cell growth is no longer regulated and cell proliferation can go on without any inhibition. The important number of cell divisions leads to the forming of tumors and then to multiploid colorectal cancer and pancreatic carcinoma. It is found inactivated in at least 50% of pancreatic cancers.[21]
Somatic mutations found in human cancers of the MH1 domain of SMAD 4 have been shown to inhibit the DNA-binding function of this domain.
SMAD 4 is also found mutated in the autosomal dominant disease juvenile polyposis syndrome (JPS). JPS is characterized by hamartomatous polyps in the gastrointestinal (GI) tract. These polyps are usually benign, however they are at greater risk of developing gastrointestinal cancers, in particular colon cancer. Around 60 mutations causing JPS have been identified. They have been linked to the production of a smaller SMAD 4, with missing domains that prevent the protein from binding to R-SMADS and forming heteromeric complexes.[4]
Mutations in SMAD4 (mostly substitutions) can cause Myhre syndrome , a rare inherited disorder characterized by mental disabilities, short stature, unusual facial features, and various bone abnormalities.[22][23]
References
- ↑ Massagué, Joan (2012). "TGFβ signalling in context". Nature Reviews Molecular Cell Biology. 13 (10): 616–630. doi:10.1038/nrm3434. ISSN 1471-0080. PMC 4027049.
- ↑ 2.0 2.1 Massagué, J. (1998). "TGF-β SIGNAL TRANSDUCTION". Annual Review of Biochemistry. 67 (1): 753–791. doi:10.1146/annurev.biochem.67.1.753. ISSN 0066-4154.
- ↑ "SMAD4 SMAD family member 4". Entrez Gene.
- ↑ 4.0 4.1 "SMAD 4". The Genetics Home Reference Website.
- ↑ Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE (Mar 1998). "Human Smad3 and Smad4 are sequence-specific transcription activators". Molecular Cell. 1 (4): 611–617. doi:10.1016/s1097-2765(00)80061-1. PMID 9660945.
- ↑ 6.0 6.1 Martin-Malpartida, Pau; Batet, Marta; Kaczmarska, Zuzanna; Freier, Regina; Gomes, Tiago; Aragón, Eric; Zou, Yilong; Wang, Qiong; Xi, Qiaoran (2017-12-12). "Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors". Nature Communications. 8 (1). doi:10.1038/s41467-017-02054-6. ISSN 2041-1723.
- ↑ Zhang, Ying; Feng, Xin-Hua; Derynck, Rik (1998). "Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription". Nature. 394 (6696): 909–913. doi:10.1038/29814. ISSN 0028-0836.
- ↑ Baburajendran, Nithya; Jauch, Ralf; Tan, Clara Yueh Zhen; Narasimhan, Kamesh; Kolatkar, Prasanna R. (2011). "Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers". Nucleic Acids Research. 39 (18): 8213–8222. doi:10.1093/nar/gkr500. ISSN 1362-4962.
- ↑ Hata, Akiko; Lo, Roger S.; Wotton, David; Lagna, Giorgio; Massagué, Joan (1997). "Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4". Nature. 388 (6637): 82–87. doi:10.1038/40424. ISSN 0028-0836.
- ↑ Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM (Mar 1995). "Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster". Genetics. 139 (3): 1347–58. PMC 1206461. PMID 7768443.
- ↑ White M (26 September 2014). "Sonic Hedgehog, DICER, and the Problem With Naming Genes". Pacific Standard.
- ↑ Roelen BA, Cohen OS, Raychowdhury MK, Chadee DN, Zhang Y, Kyriakis JM, Alessandrini AA, Lin HY (Oct 2003). "Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation". American Journal of Physiology. Cell Physiology. 285 (4): C823–30. doi:10.1152/ajpcell.00053.2003. PMID 12801888.
- ↑ Demagny H, Araki T, De Robertis EM (Oct 2014). "The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-β signaling". Cell Reports. 9 (2): 688–700. doi:10.1016/j.celrep.2014.09.020. PMID 25373906.
- ↑ de Caestecker, Mark P. (2000). "The Smad4 Activation Domain (SAD) Is a Proline-rich, p300-dependent Transcriptional Activation Domain". The Journal of Biological Chemistry. 275 (3): 2115–2122. doi:10.1074/jbc.275.3.2115. PMID 10636916.
- ↑ Demagny H, De Robertis EM (2015). "Smad4/DPC4: a Barrier against Tumor Progression driven by RTK/Ras/Erk and Wnt/GSK3 signaling". Molecular & Cellular Oncology. 3 (2): e989133. doi:10.4161/23723556.2014.989133. PMC 4905428.
- ↑ Demagny H, De Robertis EM (2015). "Point Mutations in the Tumor Suppressor Smad4/DPC4 Enhance its Phosphorylation by GSK3 and Reversibly Inactivate TGF-β Signaling". Molecular & Cellular Oncology. 3: e1025181. doi:10.1080/23723556.2015.1025181. PMC 4845174.
- ↑ Inman GJ (Feb 2005). "Linking Smads and transcriptional activation". The Biochemical Journal. 386 (Pt 1): e1–e3. doi:10.1042/bj20042133. PMC 1134782. PMID 15702493.
- ↑ Shi J, Yoshino O, Osuga Y, Nishii O, Yano T, Taketani Y (Mar 2010). "Bone morphogenetic protein 7 (BMP-7) increases the expression of follicle-stimulating hormone (FSH) receptor in human granulosa cells". Fertility and Sterility. 93 (4): 1273–9. doi:10.1016/j.fertnstert.2008.11.014. PMID 19108831.
- ↑ Pangas SA, Li X, Robertson EJ, Matzuk MM (Jun 2006). "Premature luteinization and cumulus cell defects in ovarian-specific Smad4 knockout mice". Molecular Endocrinology. 20 (6): 1406–22. doi:10.1210/me.2005-0462. PMID 16513794.
- ↑ Middlebrook BS, Eldin K, Li X, Shivasankaran S, Pangas SA (December 2009)."Smad1-Smad5 ovarian conditional knockout mice develop a disease profile similar to the juvenile form of human granulosa cell tumors", Endocrinology, 150(12):5208-17. doi: 10.1210/en.2009-0644
- ↑ Cotran RS, Kumar V, Fausto N, Robbins SL, Abbas AK (2005). Robbins and Cotran pathologic basis of disease (7th ed.). St. Louis, Mo: Elsevier Saunders. ISBN 0-7216-0187-1.
- ↑ "Growth-Mental Deficiency Syndrome of Myhre". National Organization for rare disorders.
- ↑ Caputo V, Bocchinfuso G, Castori M, Traversa A, Pizzuti A, Stella L, Grammatico P, Tartaglia M (Jul 2014). "Novel SMAD4 mutation causing Myhre syndrome". American Journal of Medical Genetics Part A. 164A (7): 1835–40. doi:10.1002/ajmg.a.36544. PMID 24715504.
Further reading
- Miyazono K (2000). "TGF-beta signaling by Smad proteins". Cytokine & Growth Factor Reviews. 11 (1–2): 15–22. doi:10.1016/S1359-6101(99)00025-8. PMID 10708949.
- Wrana JL, Attisano L (2000). "The Smad pathway". Cytokine & Growth Factor Reviews. 11 (1–2): 5–13. doi:10.1016/S1359-6101(99)00024-6. PMID 10708948.
- Verschueren K, Huylebroeck D (2000). "Remarkable versatility of Smad proteins in the nucleus of transforming growth factor-beta activated cells". Cytokine & Growth Factor Reviews. 10 (3–4): 187–99. doi:10.1016/S1359-6101(99)00012-X. PMID 10647776.
- Massagué J (1998). "TGF-beta signal transduction". Annual Review of Biochemistry. 67: 753–91. doi:10.1146/annurev.biochem.67.1.753. PMID 9759503.
- Klein-Scory S, Zapatka M, Eilert-Micus C, Hoppe S, Schwarz E, Schmiegel W, Hahn SA, Schwarte-Waldhoff I (2008). "High-level inducible Smad4-reexpression in the cervical cancer cell line C4-II is associated with a gene expression profile that predicts a preferential role of Smad4 in extracellular matrix composition". BMC Cancer. 7: 209. doi:10.1186/1471-2407-7-209. PMC 2186346. PMID 17997817.
- Kalo E, Buganim Y, Shapira KE, Besserglick H, Goldfinger N, Weisz L, Stambolsky P, Henis YI, Rotter V (Dec 2007). "Mutant p53 attenuates the SMAD-dependent transforming growth factor beta1 (TGF-beta1) signaling pathway by repressing the expression of TGF-beta receptor type II". Molecular and Cellular Biology. 27 (23): 8228–42. doi:10.1128/MCB.00374-07. PMC 2169171. PMID 17875924.
- Aretz S, Stienen D, Uhlhaas S, Stolte M, Entius MM, Loff S, Back W, Kaufmann A, Keller KM, Blaas SH, Siebert R, Vogt S, Spranger S, Holinski-Feder E, Sunde L, Propping P, Friedl W (Nov 2007). "High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome". Journal of Medical Genetics. 44 (11): 702–9. doi:10.1136/jmg.2007.052506. PMC 2752176. PMID 17873119.
- Ali S, Cohen C, Little JV, Sequeira JH, Mosunjac MB, Siddiqui MT (Oct 2007). "The utility of SMAD4 as a diagnostic immunohistochemical marker for pancreatic adenocarcinoma, and its expression in other solid tumors". Diagnostic Cytopathology. 35 (10): 644–8. doi:10.1002/dc.20715. PMID 17854080.
- Milet J, Dehais V, Bourgain C, Jouanolle AM, Mosser A, Perrin M, Morcet J, Brissot P, David V, Deugnier Y, Mosser J (Oct 2007). "Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance". American Journal of Human Genetics. 81 (4): 799–807. doi:10.1086/520001. PMC 2227929. PMID 17847004.
- Salek C, Benesova L, Zavoral M, Nosek V, Kasperova L, Ryska M, Strnad R, Traboulsi E, Minarik M (Jul 2007). "Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer". World Journal of Gastroenterology. 13 (27): 3714–20. doi:10.3748/wjg.v13.i27.3714. PMC 4250643. PMID 17659731.
- Sebestyén A, Hajdu M, Kis L, Barna G, Kopper L (Sep 2007). "Smad4-independent, PP2A-dependent apoptotic effect of exogenous transforming growth factor beta 1 in lymphoma cells". Experimental Cell Research. 313 (15): 3167–74. doi:10.1016/j.yexcr.2007.05.028. PMID 17643425.
- Martin MM, Buckenberger JA, Jiang J, Malana GE, Knoell DL, Feldman DS, Elton TS (Sep 2007). "TGF-beta1 stimulates human AT1 receptor expression in lung fibroblasts by cross talk between the Smad, p38 MAPK, JNK, and PI3K signaling pathways". American Journal of Physiology. Lung Cellular and Molecular Physiology. 293 (3): L790–9. doi:10.1152/ajplung.00099.2007. PMC 2413071. PMID 17601799.
- Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS (Sep 2007). "Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation". Molecular and Cellular Biology. 27 (17): 6068–83. doi:10.1128/MCB.00664-07. PMC 1952153. PMID 17591695.
- Grijelmo C, Rodrigue C, Svrcek M, Bruyneel E, Hendrix A, de Wever O, Gespach C (Aug 2007). "Proinvasive activity of BMP-7 through SMAD4/src-independent and ERK/Rac/JNK-dependent signaling pathways in colon cancer cells". Cellular Signalling. 19 (8): 1722–32. doi:10.1016/j.cellsig.2007.03.008. PMID 17478078.
- Sonegawa H, Nukui T, Li DW, Takaishi M, Sakaguchi M, Huh NH (Jul 2007). "Involvement of deterioration in S100C/A11-mediated pathway in resistance of human squamous cancer cell lines to TGFbeta-induced growth suppression". Journal of Molecular Medicine. 85 (7): 753–62. doi:10.1007/s00109-007-0180-7. PMID 17476473.
- Sheikh AA, Vimalachandran D, Thompson CC, Jenkins RE, Nedjadi T, Shekouh A, Campbell F, Dodson A, Prime W, Crnogorac-Jurcevic T, Lemoine NR, Costello E (Jun 2007). "The expression of S100A8 in pancreatic cancer-associated monocytes is associated with the Smad4 status of pancreatic cancer cells". Proteomics. 7 (11): 1929–40. doi:10.1002/pmic.200700072. PMID 17469085.
- Popović Hadzija M, Korolija M, Jakić Razumović J, Pavković P, Hadzija M, Kapitanović S (Apr 2007). "K-ras and Dpc4 mutations in chronic pancreatitis: case series". Croatian Medical Journal. 48 (2): 218–24. PMC 2080529. PMID 17436386.
- Losi L, Bouzourene H, Benhattar J (May 2007). "Loss of Smad4 expression predicts liver metastasis in human colorectal cancer". Oncology Reports. 17 (5): 1095–9. doi:10.3892/or.17.5.1095. PMID 17390050.
- Karlsson G, Blank U, Moody JL, Ehinger M, Singbrant S, Deng CX, Karlsson S (Mar 2007). "Smad4 is critical for self-renewal of hematopoietic stem cells". The Journal of Experimental Medicine. 204 (3): 467–74. doi:10.1084/jem.20060465. PMC 2137898. PMID 17353364.
- Takano S, Kanai F, Jazag A, Ijichi H, Yao J, Ogawa H, Enomoto N, Omata M, Nakao A (Mar 2007). "Smad4 is essential for down-regulation of E-cadherin induced by TGF-beta in pancreatic cancer cell line PANC-1". Journal of Biochemistry. 141 (3): 345–51. doi:10.1093/jb/mvm039. PMID 17301079.
External links
- GeneReviews/NCBI/NIH/UW entry on Hereditary Hemorrhagic Telangiectasia
- GeneReviews/NCBI/NIH/UW entry on Juvenile Polyposis Syndrome
- SMAD4 gene variant database