Cancer pathophysiology
|
Cancer Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Cancer pathophysiology On the Web |
|
American Roentgen Ray Society Images of Cancer pathophysiology |
|
Risk calculators and risk factors for Cancer pathophysiology |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview

Cancer is fundamentally a disease of regulation of tissue growth. In order for a normal cell to transform into a cancer cell, genes which regulate cell growth and differentiation must be altered. Genetic changes can occur at many levels, from gain or loss of entire chromosomes to a mutation affecting a single DNA nucleotide. There are two broad categories of genes which are affected by these changes. Oncogenes may be normal genes which are expressed at inappropriately high levels, or altered genes which have novel properties. In either case, expression of these genes promotes the malignant phenotype of cancer cells. Tumor suppressor genes are genes which inhibit cell division, survival, or other properties of cancer cells. Tumor suppressor genes are often disabled by cancer-promoting genetic changes. Typically, changes in many genes are required to transform a normal cell into a cancer cell.
There is a diverse classification scheme for the various genomic changes which may contribute to the generation of cancer cells. Most of these changes are mutations, or changes in the nucleotide sequence of genomic DNA. Aneuploidy, the presence of an abnormal number of chromosomes, is one genomic change which is not a mutation, and may involve either gain or loss of one or more chromosomes through errors in mitosis.
Large-scale mutations involve the deletion or gain of a portion of a chromosome. Genomic amplification occurs when a cell gains many copies (often 20 or more) of a small chromosomal locus, usually containing one or more oncogenes and adjacent genetic material. Translocation occurs when two separate chromosomal regions become abnormally fused, often at a characteristic location. A well-known example of this is the Philadelphia chromosome, or translocation of chromosomes 9 and 22, which occurs in chronic myelogenous leukemia, and results in production of the BCR-abl fusion protein, an oncogenic tyrosine kinase.
Small-scale mutations include point mutations, deletions, and insertions, which may occur in the promoter of a gene and affect its expression, or may occur in the gene's coding sequence and alter the function or stability of its protein product. Disruption of a single gene may also result from integration of genomic material from a DNA virus or retrovirus, and such an event may also result in the expression of viral oncogenes in the affected cell and its descendants.
Pathophysiology
Epigenetics
Epigenetics is the study of the regulation of gene expression through chemical, non-mutational changes in DNA structure. The theory of epigenetics in cancer pathogenesis is that non-mutational changes to DNA can lead to alterations in gene expression. Normally, oncogenes are silent, for example, because of DNA methylation. Loss of that methylation can induce the aberrant expression of oncogenes, leading to cancer pathogenesis. Known mechanisms of epigenetic change include DNA methylation, and methylation or acetylation of histone proteins bound to chromosomal DNA at specific locations. Classes of medications, known as HDAC inhibitors and DNA methyltransferase inhibitors, can re-regulate the epigenetic signaling in the cancer cell.
Oncogenes
Oncogenes promote cell growth through a variety of ways. Many can produce hormones, a "chemical messenger" between cells which encourage mitosis, the effect of which depends on the signal transduction of the receiving tissue or cells. In other words, when a hormone receptor on a recipient cell is stimulated, the signal is conducted from the surface of the cell to the cell nucleus to effect some change in gene transcription regulation at the nuclear level. Some oncogenes are part of the signal transduction system itself, or the signal receptors in cells and tissues themselves, thus controlling the sensitivity to such hormones. Oncogenes often produce mitogens, or are involved in transcription of DNA in protein synthesis, which creates the proteins and enzymes responsible for producing the products and biochemicals cells use and interact with.
Mutations in proto-oncogenes, which are the normally quiescent counterparts of oncogenes, can modify their expression and function, increasing the amount or activity of the product protein. When this happens, the proto-oncogenes become oncogenes, and this transition upsets the normal balance of cell cycle regulation in the cell, making uncontrolled growth possible. The chance of cancer cannot be reduced by removing proto-oncogenes from the genome, even if this were possible, as they are critical for growth, repair and homeostasis of the organism. It is only when they become mutated that the signals for growth become excessive.
One of the first oncogenes to be defined in cancer research is the ras oncogene. Mutations in the Ras family of proto-oncogenes (comprising H-Ras, N-Ras and K-Ras) are very common, being found in 20% to 30% of all human tumours.[1] Ras was originally identified in the Harvey sarcoma virus genome, and researchers were surprised that not only was this gene present in the human genome but that, when ligated to a stimulating control element, could induce cancers in cell line cultures.[2]
Tumor Suppressor Genes
Tumor suppressor genes code for anti-proliferation signals and proteins that suppress mitosis and cell growth. Generally, tumor suppressors are transcription factors that are activated by cellular stress or DNA damage. Often DNA damage will cause the presence of free-floating genetic material as well as other signs, and will trigger enzymes and pathways which lead to the activation of tumor suppressor genes. The functions of such genes is to arrest the progression of the cell cycle in order to carry out DNA repair, preventing mutations from being passed on to daughter cells. The p53 protein, one of the most important studied tumor suppressor genes, is a transcription factor activated by many cellular stressors including hypoxia and ultraviolet radiation damage.
Despite nearly half of all cancers possibly involving alterations in p53, its tumor suppressor function is poorly understood. p53 clearly has two functions: one a nuclear role as a transcription factor, and the other a cytoplasmic role in regulating the cell cycle, cell division, and apoptosis.
The Warburg hypothesis is the preferential use of glycolysis for energy to sustain cancer growth. p53 has been shown to regulate the shift from the respiratory to the glycolytic pathway.[3]
However, a mutation can damage the tumor suppressor gene itself, or the signal pathway which activates it, "switching it off". The invariable consequence of this is that DNA repair is hindered or inhibited: DNA damage accumulates without repair, inevitably leading to cancer.
Mutations of tumor suppressor genes that occur in germline cells are passed along to offspring, and increase the likelihood for cancer diagnoses in subsequent generations. Members of these families have increased incidence and decreased latency of multiple tumors. The tumor types are typical for each type of tumor suppressor gene mutation, with some mutations causing particular cancers, and other mutations causing others. The mode of inheritance of mutant tumor suppressors is that an affected member inherits a defective copy from one parent, and a normal copy from the other. For instance, individuals who inherit one mutant p53 allele (and are therefore heterozygous for mutated p53) can develop melanomas and pancreatic cancer, known as Li-Fraumeni syndrome. Other inherited tumor suppressor gene syndromes include Rb mutations, linked to retinoblastoma, and APC gene mutations, linked to adenopolyposis colon cancer. Adenopolyposis colon cancer is associated with thousands of polyps in colon while young, leading to colon cancer at a relatively early age. Finally, inherited mutations in BRCA1 and BRCA2 lead to early onset of breast cancer.
Development of cancer was proposed in 1971 to depend on at least two mutational events. In what became known as the Knudson two-hit hypothesis, an inherited, germ-line mutation in a tumor suppressor gene would only cause cancer if another mutation event occurred later in the organism's life, inactivating the other allele of that tumor suppressor gene.[4]
Usually, oncogenes are dominant, as they contain gain-of-function mutations, while mutated tumor suppressors are recessive, as they contain loss-of-function mutations. Each cell has two copies of the same gene, one from each parent, and under most cases gain of function mutations in just one copy of a particular proto-oncogene is enough to make that gene a true oncogene. On the other hand, loss of function mutations need to happen in both copies of a tumor suppressor gene to render that gene completely non-functional. However, cases exist in which one mutated copy of a tumor suppressor gene can render the other, wild-type copy non-functional. This phenomenon is called the dominant negative effect and is observed in many p53 mutations.
Knudson’s two hit model has recently been challenged by several investigators. Inactivation of one allele of some tumor suppressor genes is sufficient to cause tumors. This phenomenon is called haploinsufficiency and has been demonstrated by a number of experimental approaches. Tumors caused by haploinsufficiency usually have a later age of onset when compared with those by a two hit process.[5]
Cancer Cell Biology
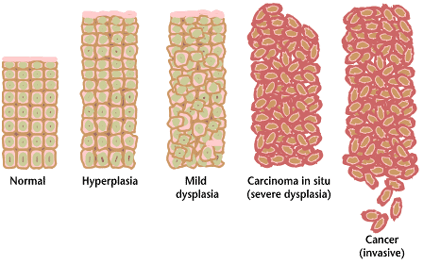
Often, the multiple genetic changes which result in cancer may take many years to accumulate. During this time, the biological behavior of the pre-malignant cells slowly change from the properties of normal cells to cancer-like properties. Pre-malignant tissue can have a distinctive appearance under the microscope. Among the distinguishing traits are an increased number of dividing cells, variation in nuclear size and shape, variation in cell size and shape, loss of specialized cell features, and loss of normal tissue organization. Dysplasia is an abnormal type of excessive cell proliferation characterized by loss of normal tissue arrangement and cell structure in pre-malignant cells. These early neoplastic changes must be distinguished from hyperplasia, a reversible increase in cell division caused by an external stimulus, such as a hormonal imbalance or chronic irritation.
The most severe cases of dysplasia are referred to as "carcinoma in situ." In Latin, the term "in situ" means "in place", so carcinoma in situ refers to an uncontrolled growth of cells that remains in the original location and has not shown invasion into other tissues. Nevertheless, carcinoma in situ may develop into an invasive malignancy and is usually removed surgically, if possible.
Clonal Evolution
The process of malignancy can be explained from an evolutionary perspective. Millions of years of biological evolution insure that the cellular metabolic changes that enable cancer to grow occur only very rarely. Most changes in cellular metabolism that allow cells to grow in a disorderly fashion lead to cell death. Cancer cells undergo a process analogous to natural selection, in that the few cells with new genetic changes that enhance their survival continue to multiply, and soon come to dominate the growing tumor, as cells with less favorable genetic change are outcompeted. This process is called clonal evolution. Tumors often continue to evolve in response to chemotherapy treatments, and on occasion aberrant cells may acquire resistance to particular anti-cancer pharmaceuticals.
Biological Properties of Cancer Cells
In a 2000 article by Hanahan and Weinberg, the biological properties of malignant tumor cells were summarized as follows:[6]
- Acquisition of self-sufficiency in growth signals, leading to unchecked growth.
- Loss of sensitivity to anti-growth signals, also leading to unchecked growth.
- Loss of capacity for apoptosis, in order to allow growth despite genetic errors and external anti-growth signals.
- Loss of capacity for senescence, leading to limitless replicative potential (immortality)
- Acquisition of sustained angiogenesis, allowing the tumor to grow beyond the limitations of passive nutrient diffusion.
- Acquisition of ability to invade neighbouring tissues, the defining property of invasive carcinoma.
- Acquisition of ability to build metastases at distant sites, the classical property of malignant tumors (carcinomas or others).
The completion of these multiple steps would be a very rare event without :
- Loss of capacity to repair genetic errors, leading to an increased mutation rate (genomic instability), thus accelerating all the other changes.
These biological changes are classical in carcinomas; other malignant tumor may not need all to achieve them all. For example, tissue invasion and displacement to distant sites are normal properties of leukocytes; these steps are not needed in the development of Leukemia. The different steps do not necessarily represent individual mutations. For example, inactivation of a single gene, coding for the P53 protein, will cause genomic instability, evasion of apoptosis and increased angiogenesis.
References
- ↑ Bos J (1989). "ras oncogenes in human cancer: a review". Cancer Res. 49 (17): 4682–9. PMID 2547513.
- ↑ Chang EH, Furth ME, Scolnick EM, Lowy DR (1982). "Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus". Nature. 297 (5866): 479–83. PMID 6283358.
- ↑ Matoba S, Kang J, Patino W, Wragg A, Boehm M, Gavrilova O, Hurley P, Bunz F, Hwang P (2006). "p53 regulates mitochondrial respiration". Science. 312 (5780): 1650–3. PMID 16728594.
- ↑ Knudson A (1971). "Mutation and cancer: statistical study of retinoblastoma". Proc Natl Acad Sci U S A. 68 (4): 820–3. PMID 5279523.
- ↑ Fodde R, Smits R (2002). "Cancer biology. A matter of dosage". Science. 298 (5594): 761–3. PMID 12399571.
- ↑ Hanahan D, Weinberg RA (2000). "The hallmarks of cancer". Cell. 100 (1): 57–70. PMID 10647931.