Adenosine deaminase
| Adenosine deaminase | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||
| |||||||||||||
| Identifiers | |||||||||||||
| Symbols | ADA ; | ||||||||||||
| External IDs | Template:OMIM5 Template:MGI HomoloGene: 37249 | ||||||||||||
| |||||||||||||
| RNA expression pattern | |||||||||||||
 | |||||||||||||
 | |||||||||||||
| More reference expression data | |||||||||||||
| Orthologs | |||||||||||||
| Template:GNF Ortholog box | |||||||||||||
| Species | Human | Mouse | |||||||||||
| Entrez | n/a | n/a | |||||||||||
| Ensembl | n/a | n/a | |||||||||||
| UniProt | n/a | n/a | |||||||||||
| RefSeq (mRNA) | n/a | n/a | |||||||||||
| RefSeq (protein) | n/a | n/a | |||||||||||
| Location (UCSC) | n/a | n/a | |||||||||||
| PubMed search | n/a | n/a | |||||||||||
Adenosine deaminase (also known as ADA) is an enzyme (EC 3.5.4.4) involved in purine metabolism. It is needed for the breakdown of adenosine from food and for the turnover of nucleic acids in tissues.
Reactions
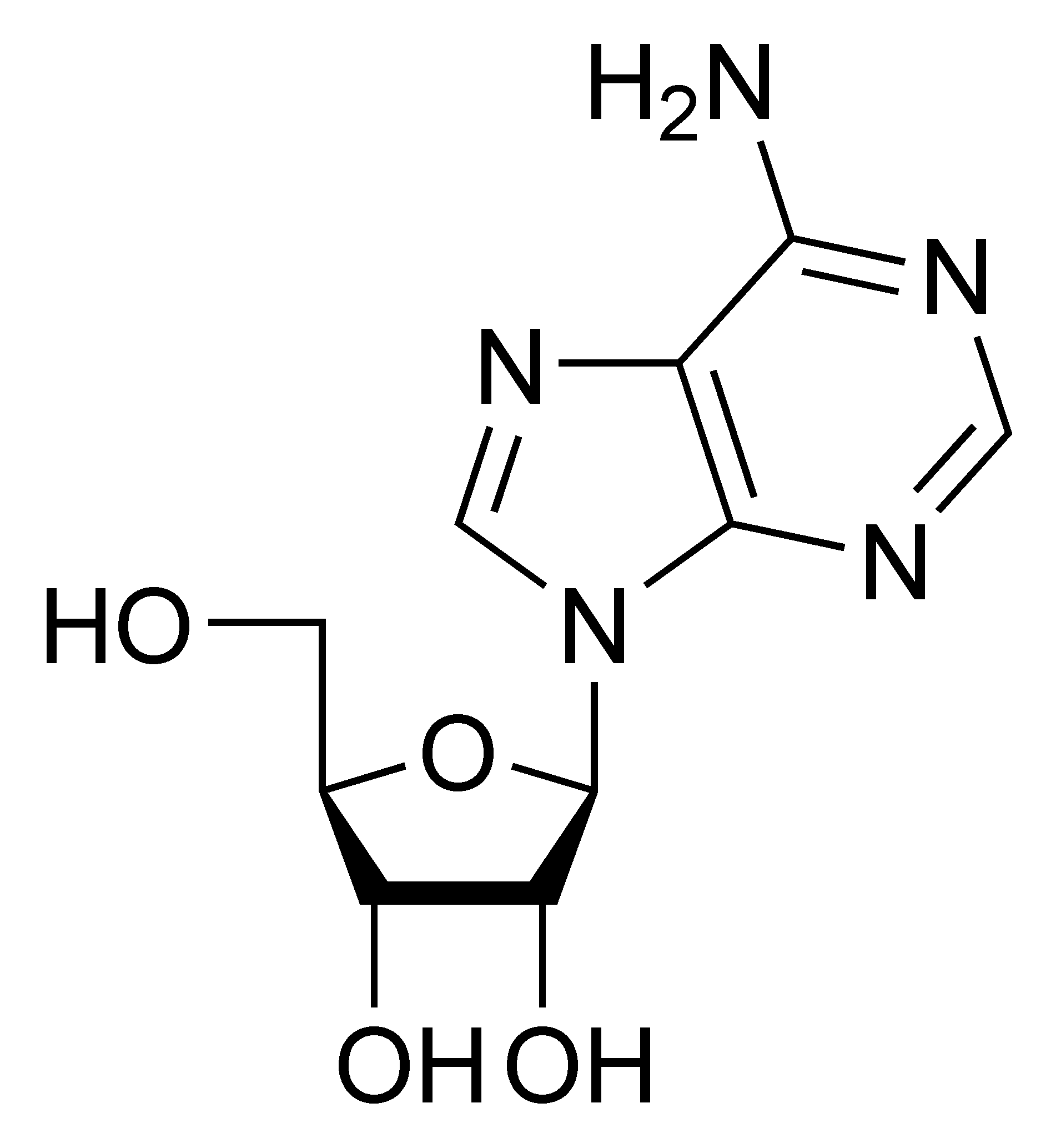
ADA irreversibly deaminates adenosine, converting it to the related nucleoside inosine by the removal of an amino group.
-
Inosine (observe missing nitrogen in upper left)
Inosine can then be deribosylated (removed from ribose) by another enzyme called purine nucleoside phosphorylase (PNP), converting it to hypoxanthine.
Pathology
Mutations in the gene for adenosine deaminase causing it to not be expressed are one cause of severe combined immunodeficiency (SCID).[1]
Mutations causing it to be overexpressed are one cause of hemolytic anemia.[2]
There is some evidence that a different allelle (ADA2) may lead to autism.[3]
Isoforms
There are 2 isoforms of ADA: ADA1 and ADA2.
- ADA1 is found in most body cells, particularly lymphocytes and macrophages, where it is present not only in the cytosol and nucleus but also as the ecto- form on the cell membrane attached to dipeptidyl peptidase-4 (aka, CD26).
- ADA2 was first identified in human thymus. It was subsequently found in other tissues including the macrophage where it co-exists with ADA1. The two isoforms regulate the ratio of adenosine to deoxyadenosine potentiating the killing of parasites.
- ADAT (ADAT1, ADAT2, ADAT3) is a tRNA-specific ADA, changing the tRNA to allow for a wobble base pairing.
Clinical significance
ADA2 is the predominant form present in human blood plasma and is increased in many diseases, particularly those associated with the immune system: for example rheumatoid arthritis, psoriasis and sarcoidosis. The plasma AD2 isoform is also increased in most cancers.
Total plasma ADA can be measured using high performance liquid chromatography, enzymatic or colorimetric techniques. Perhaps the simplest system is the measurement of the ammonia released from adenosine when broken down to inosine. After incubation of plasma with a buffered solution of adenosine the ammonia is reacted with a Berthelot reagent to form a blue colour which is proportionate to the amount of enzyme activity. To measure ADA2, erythro-9-(2-hydroxy-3-nonyl) adenine (EHNA) is added prior to incubation so as to inhibit the enzymatic acivity of ADA1. It is the absence of ADA1 that causes SCID.
See also
- Adenosine deaminase deficiency
- ADAR, a human gene encoding a RNA-specific adenosine deaminase
References
- ↑ Sanchez JJ, Monaghan G, Børsting C, Norbury G, Morling N, Gaspar HB (2007). "Carrier frequency of a nonsense mutation in the adenosine deaminase (ADA) gene implies a high incidence of ADA-deficient severe combined immunodeficiency (SCID) in Somalia and a single, common haplotype indicates common ancestry". Ann. Hum. Genet. 71 (Pt 3): 336–47. doi:10.1111/j.1469-1809.2006.00338.x. PMID 17181544.
- ↑ Chottiner EG, Cloft HJ, Tartaglia AP, Mitchell BS (1987). "Elevated adenosine deaminase activity and hereditary hemolytic anemia. Evidence for abnormal translational control of protein synthesis". J. Clin. Invest. 79 (3): 1001–5. PMID 3029177.
- ↑ Persico AM, Militerni R, Bravaccio C; et al. (2000). "Adenosine deaminase alleles and autistic disorder: case-control and family-based association studies". Am. J. Med. Genet. 96 (6): 784–90. PMID 11121182.
- ↑ Keegan LP, Leroy A, Sproul D, O'Connell MA (2004). "Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes". Genome Biol. 5 (2): 209. doi:10.1186/gb-2004-5-2-209. PMID 14759252.
Further reading
- da Cunha JG (1992). "[Adenosine deaminase. A pluridisciplinary enzyme]". Acta médica portuguesa. 4 (6): 315–23. PMID 1807098.
- Franco R, Casadó V, Ciruela F; et al. (1997). "Cell surface adenosine deaminase: much more than an ectoenzyme". Prog. Neurobiol. 52 (4): 283–94. PMID 9247966.
- Valenzuela A, Blanco J, Callebaut C; et al. (1997). "HIV-1 envelope gp120 and viral particles block adenosine deaminase binding to human CD26". Adv. Exp. Med. Biol. 421: 185–92. PMID 9330696.
- Moriwaki Y, Yamamoto T, Higashino K (1999). "Enzymes involved in purine metabolism--a review of histochemical localization and functional implications". Histol. Histopathol. 14 (4): 1321–40. PMID 10506947.
- Hirschhorn R (1993). "Identification of two new missense mutations (R156C and S291L) in two ADA- SCID patients unusual for response to therapy with partial exchange transfusions". Hum. Mutat. 1 (2): 166–8. doi:10.1002/humu.1380010214. PMID 1284479.
- Berkvens TM, van Ormondt H, Gerritsen EJ; et al. (1990). "Identical 3250-bp deletion between two AluI repeats in the ADA genes of unrelated ADA-SCID patients". Genomics. 7 (4): 486–90. PMID 1696926.
- Aran JM, Colomer D, Matutes E; et al. (1991). "Presence of adenosine deaminase on the surface of mononuclear blood cells: immunochemical localization using light and electron microscopy". J. Histochem. Cytochem. 39 (8): 1001–8. PMID 1856451.
- Bielat K, Tritsch GL (1989). "Ecto-enzyme activity of human erythrocyte adenosine deaminase". Mol. Cell. Biochem. 86 (2): 135–42. PMID 2770711.
- Hirschhorn R, Tzall S, Ellenbogen A, Orkin SH (1989). "Identification of a point mutation resulting in a heat-labile adenosine deaminase (ADA) in two unrelated children with partial ADA deficiency". J. Clin. Invest. 83 (2): 497–501. PMID 2783588.
- Murray JL, Perez-Soler R, Bywaters D, Hersh EM (1986). "Decreased adenosine deaminase (ADA) and 5'nucleotidase (5NT) activity in peripheral blood T cells in Hodgkin disease". Am. J. Hematol. 21 (1): 57–66. PMID 3010705.
- Wiginton DA, Kaplan DJ, States JC; et al. (1987). "Complete sequence and structure of the gene for human adenosine deaminase". Biochemistry. 25 (25): 8234–44. PMID 3028473.
- Akeson AL, Wiginton DA, Dusing MR; et al. (1988). "Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts". J. Biol. Chem. 263 (31): 16291–6. PMID 3182793.
- Glader BE, Backer K (1988). "Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases". Br. J. Haematol. 68 (2): 165–8. PMID 3348976.
- Petersen MB, Tranebjaerg L, Tommerup N; et al. (1987). "New assignment of the adenosine deaminase gene locus to chromosome 20q13 X 11 by study of a patient with interstitial deletion 20q". J. Med. Genet. 24 (2): 93–6. PMID 3560174.
- Orkin SH, Goff SC, Kelley WN, Daddona PE (1985). "Transient expression of human adenosine deaminase cDNAs: identification of a nonfunctional clone resulting from a single amino acid substitution". Mol. Cell. Biol. 5 (4): 762–7. PMID 3838797.
- Valerio D, Duyvesteyn MG, Dekker BM; et al. (1985). "Adenosine deaminase: characterization and expression of a gene with a remarkable promoter". EMBO J. 4 (2): 437–43. PMID 3839456.
- Bonthron DT, Markham AF, Ginsburg D, Orkin SH (1985). "Identification of a point mutation in the adenosine deaminase gene responsible for immunodeficiency". J. Clin. Invest. 76 (2): 894–7. PMID 3839802.
- Daddona PE, Shewach DS, Kelley WN; et al. (1984). "Human adenosine deaminase. cDNA and complete primary amino acid sequence". J. Biol. Chem. 259 (19): 12101–6. PMID 6090454.
- Valerio D, Duyvesteyn MG, Meera Khan P; et al. (1984). "Isolation of cDNA clones for human adenosine deaminase". Gene. 25 (2–3): 231–40. PMID 6198240.
| Stub icon | This hydrolase article is a stub. You can help Wikipedia by expanding it. |