WBR0121
| Author | [[PageAuthor::William J Gibson (reviewed by Rim Halaby, M.D. [1])]] |
|---|---|
| Exam Type | ExamType::USMLE Step 1 |
| Main Category | MainCategory::Genetics |
| Sub Category | SubCategory::Reproductive, SubCategory::General Principles |
| Prompt | [[Prompt::A 56-year-old woman presents to her primary care physician complaining of abdominal pain, bloating, and early satiety. She explains that these symptoms have been increasing in severity over the past several weeks. The physician notes mild tenderness with palpation on abdominal exam. The patient is then referred to an OB/GYN specialist where a pelvic exam is performed. Pelvic exam reveals a large left adnexal mass. Laboratory studies are significant for elevated CA-125. Abdominopelvic CT scan reveals a large left ovarian solid mass that extends to the omentum. The patient undergoes total abdominal hysterectomy and bilateral bilateral salpingo-oophorectomy (TAH-BSO). After resection, the pathologist makes the diagnosis of high-grade serous ovarian adenocarcinoma. Molecular analysis demonstrates the following sequencing results of the TP53 gene:
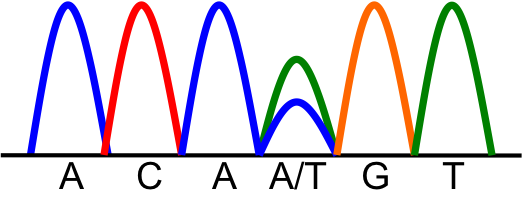 Which of the following is the most appropriate term for the molecular effect of this mutation?]] |
| Answer A | AnswerA::Loss of heterozygosity |
| Answer A Explanation | AnswerAExp::While loss of heterozygosity is a term commonly used with tumor suppressors, the mutation in this case is heterozygous. |
| Answer B | AnswerB::Dominant negative mutation |
| Answer B Explanation | AnswerBExp::A dominant negative mutation refers to a mutation whose gene product negates the function of its normal, wild-type counterpart. |
| Answer C | AnswerC::Epistasis |
| Answer C Explanation | [[AnswerCExp::Epistasis is a phenomenon in which the expression of one gene depends on the presence of other "modifier genes(s)". Both genes interact to control a single phenotype.]] |
| Answer D | AnswerD::Haploinsufficiency |
| Answer D Explanation | [[AnswerDExp::While mutation of one allele causes disease, the molecular mechanism of the mutation observed in this vignette is not simply attributed to decreased function of the monomeric gene product. In this case, the mutant protein interferes with the function of another wild-type gene]] |
| Answer E | AnswerE::Loss-of-function |
| Answer E Explanation | AnswerEExp::While the observed mutation may cause the protein to lose some of its biological function, it does so in a dominant negative manner. Loss-of-function mutations typically act in a recessive manner at the molecular level. |
| Right Answer | RightAnswer::B |
| Explanation | [[Explanation::p53 is a transcriprtional activator that controls cell fate and initiates cell cycle arrest, senescence, and apoptosis. Mutations of p53 usually affect the core domain and exhibit dominant negative effect of the mutant allele on the wild-type allele. In this vignette, the mutation demonstrates a heterozygous mutation, meaning that one copy of the TP53 gene is mutated and the other is not. A heterozygous mutation which leads to a phenotype (in this cas cancer) is an example of a dominant negative mutation. On the molecular level, the p53 protein normally forms a homotetramer, a hollow skewed cube that binds to specific DNA regions at the level of p53 core domains.
Unlike mutations of other tumor suppressors, a mutation of a single p53 allele leads to a significant loss-of-function effect due to the dominant negative effect of the mutant allele that disrupts the effect of the normal wild-type allele. One hypothesis suggests that when one mutant TP53 allele joins the wild-type homotetramer, the homotetramer is heterotetramerized; and wild-type p53 function loses its conformation and is inactivated. Thus, one mutated allele causes the p53 protein to lose approximately 15/16 of its biological function. On the other hand, a deletion of p53 would cause a 50% decrease in its biological function. For this reason, p53 is more likely to be mutated than deleted. Dominant mutant p53 proteins are notoriously over-expressed in ovarian cancers, where cross-talk between the wild-type p53 and the dominant mutant p53 protein has compromised all attempts of gene therapy and efforts that target p53 in these cancers. Willis A, Jung EJ, Wakefield T, et al. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes Oncogene. 2004;23:2330-2338 First Aid 2014 page 76]] |
| Approved | Approved::Yes |
| Keyword | WBRKeyword::Mutation, WBRKeyword::Genetics, WBRKeyword::Dominant, WBRKeyword::Negative, WBRKeyword::Dominant negative, WBRKeyword::Homotetramer, WBRKeyword::Heterotetramer, WBRKeyword::Ovarian, WBRKeyword::Cancer, WBRKeyword::Adenocarcinoma, WBRKeyword::Adnexal, WBRKeyword::Mass, WBRKeyword::Adnexal, WBRKeyword::mass, WBRKeyword::OB/GYN, WBRKeyword::Haploinsufficiency, WBRKeyword::Epistatis, WBRKeyword::Epigenetics, WBRKeyword::Loss of function, WBRKeyword::Loss of heterozygosity, WBRKeyword::Mutation, WBRKeyword::Tumor suppressor gene, WBRKeyword::Tumor suppressor, WBRKeyword::Apoptosis, WBRKeyword::Cell cycle arrest, WBRKeyword::Cell cycle, WBRKeyword::p53, WBRKeyword::TP53 |
| Linked Question | Linked:: |
| Order in Linked Questions | LinkedOrder:: |