Rotavirus
| style="background:#Template:Taxobox colour;"|Rotavirus |
|---|
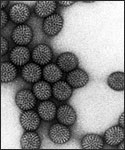 Electron micrograph of Rotaviruses.
|
| style="background:#Template:Taxobox colour;" | Scientific classification |
|
|
|
Rotavirus infection Microchapters |
|
Diagnosis |
|---|
|
Treatment |
|
Case Studies |
|
Rotavirus On the Web |
|
American Roentgen Ray Society Images of Rotavirus |
Editor-In-Chief: C. Michael Gibson, M.S., M.D. [1]
Overview
Rotaviruses are a genus of viruses belonging to the Reoviridae family. Seven major groups have been identified, three of which (groups A, B, and C) infect humans, with group A being the most common and widespread one. They cause vomiting and diarrhea and are the most common cause of severe diarrhea in children, killing about 600,000 children every year in developing countries (as of 2005). New vaccines have been shown to be safe and effective in 2006 [2].
Microbiology
 |
 |
 |
Structure
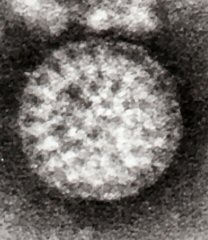
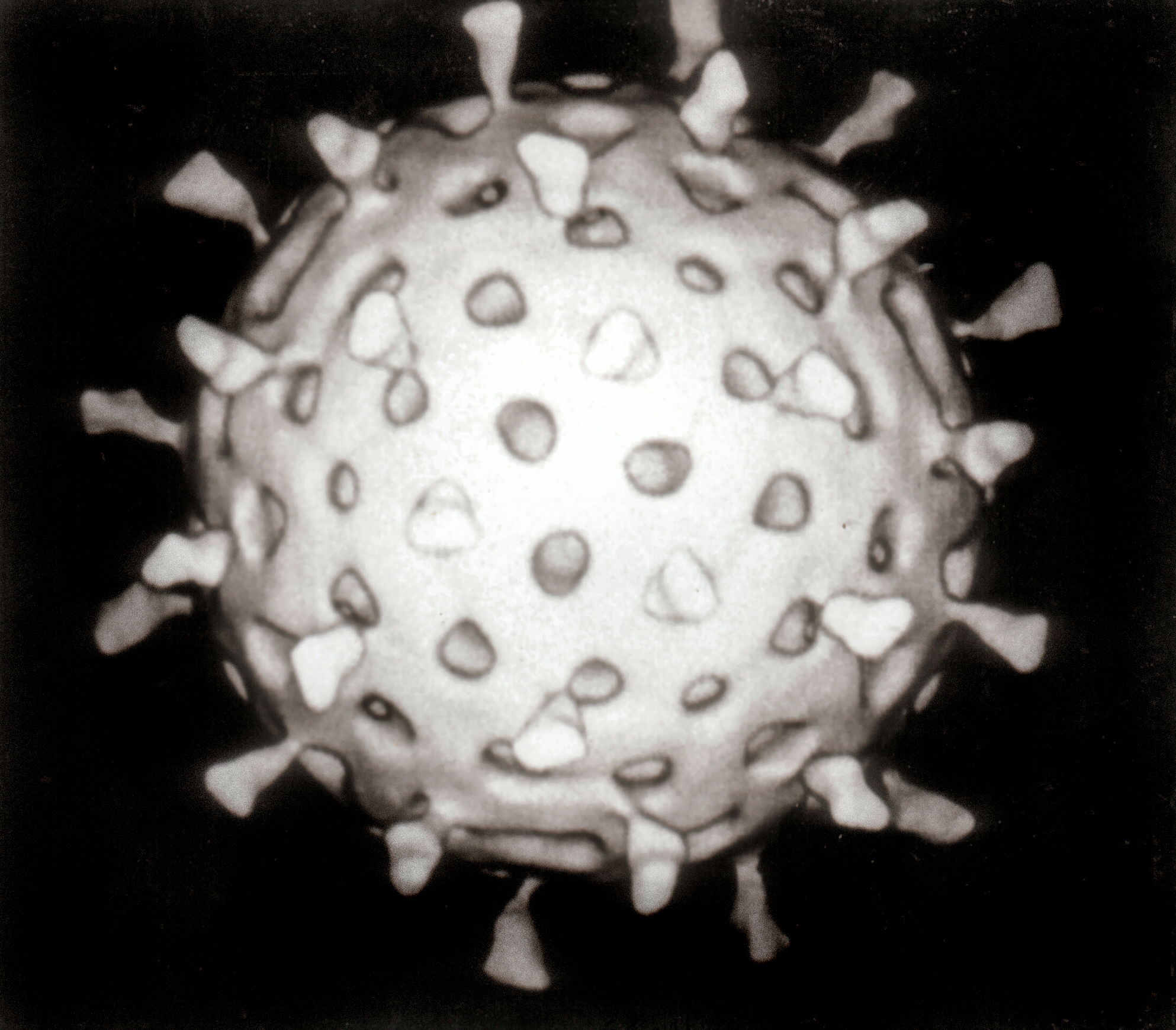
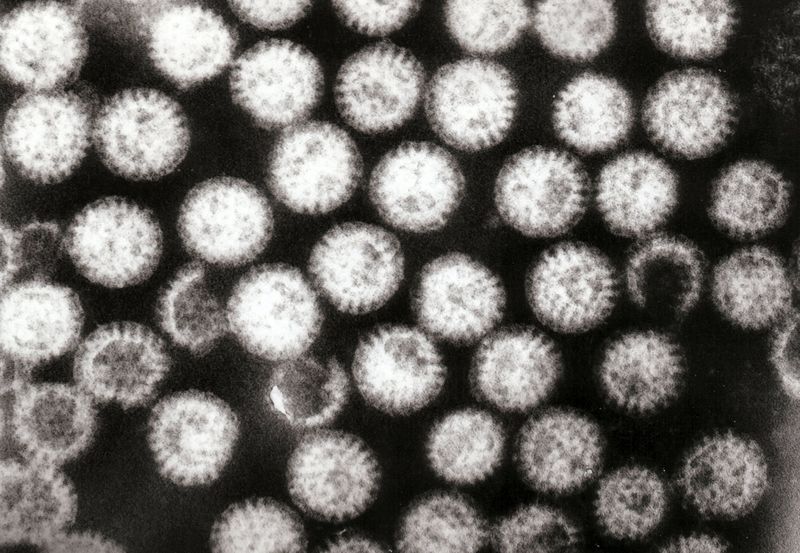
Rotaviruses have a genome consisting of 11 double-stranded RNA segments surrounded by a distinctive three-layered icosahedral protein capsid. The first layer is formed by the protein VP2, with each vertex having a copy of the proteins VP1 and VP3. The second layer is formed by the protein VP6. The outermost protein layer is composed of the structural glycoprotein VP7 and the spike protein VP4. Viral particles are up to 70nm in diameter and have a buoyant density of 1.36 g/ml in CsCl. By negative staining electron microscopy they resemble 'wheels' from which they derive their name (rota is Latin for wheel).
Cell Infection
Rotaviruses tend to affect gastrointestinal epithelial cells that are at the tip of the villus. Their triple protein coats make them very resistant to the normally prohibitive pH of the stomach, and also digestive enzymes (lipases and proteases) in the gastrointestinal tract.
When they infect a cell, they are ingested by the cell in endocytosis in a vesicle known as an endosome. Proteins in the third layer (VP7 and the VP4 spike) disrupt the membrane of the endosome, creating a difference in the Ca2+ concentration. This facilitates the breakdown of VP7 trimers into single protein subunits, leaving the VP2 and VP6 coats around the viral dsRNA, forming a double-layer particle (DLP).
While the eleven dsRNA strands are still within the protection of the two protein shells, RNA-dependent RNA polymerase creates viral mRNA transcripts of the double-stranded viral genome. This is more easily done within the environment in the "core" of the virus than in the host cell's aqueous environment, which significantly slows down the detachment of the two RNA strands to begin mRNA synthesis. Encapsidation of the viral RNA may also serve to evade host immune responses that are triggered by the presence of double-stranded RNA.
During the infection, rotavirus produces mRNA to support both protein translation and genome replication. Most of the rotavirus proteins accumulate in structures known as viroplasms, where the RNA is replicated and the DLPs are assembled. Viroplasms are electron-dense, perinuclear, punctate structures found as early as 2 hours after virus infection. Viroplasms are viral factories and are thought to be formed by two viral non-structural proteins, NSP5 and NSP2. Expression of certain forms of NSP5, especially one that is tagged at the NH2-terminus, results in the formation of viroplasms. Inhibition of NSP5 using intrabodies or RNA interference results in a profound decrease in rotavirus replication. The DLPs can migrate to the endoplasmic reticulum where they obtain their third, outer layer (formed by VP7 and VP4).
Transmission and associated foods
Rotaviruses are transmitted by the fecal-oral route. Person-to-person spread through contaminated hands is probably the most important means by which rotaviruses are transmitted in close communities such as pediatric and geriatric wards, day care centers and family homes.
Infected food handlers may contaminate foods that require handling and no further cooking, such as salads, fruits, and hors d'oeuvres. Rotaviruses are quite stable in the environment and have been found in estuary samples at levels as high as 1-5 infectious particles/gal. Sanitary measures adequate for bacteria and parasites seem to be ineffective in endemic control of rotavirus, as similar incidence of rotavirus infection is observed in countries with both high and low health standards.
The virus has not been isolated from any food associated with an outbreak, and no satisfactory method is available for routine analysis of food. However, it should be possible to apply procedures that have been used to detect the virus in water and in clinical specimens, of which reverse transcription (RT)-PCR amplification is the most sensitive method to food analysis.
Sources
- The Bad Bug Book by the U.S. Food & Drug Administration, 1992.
- New vaccines for diarrhoea virus shown effective, New Scientist, 5 January 2006
- Diarrhoea vaccines prove their mettle, news@nature.com, 5 January 2006
See also
External links
- CDC Viral Gastroenteritis FAQs: Center for Disease Control and Prevention of Food Illness Fact Sheet
- Loci index for genome Rotavirus: Available from the GenBank Taxonomy database, which contains the names of all organisms that are represented in the genetic databases with at least one nucleotide or protein sequence.
- Encyclopedia of Children's Health: Rotavirus Infections: Description, causes and symptoms, diagnosis, treatment, prognosis, prevention, parental concerns, and resources.
- Rotavirus, Emerging Infectious Diseases, Vol 4 No 4, Oct 1998.
- PATH's Rotavirus Vaccine Program
- Rotavirus disease and vaccine resources