Polyhistidine-tag
Please Take Over This Page and Apply to be Editor-In-Chief for this topic: There can be one or more than one Editor-In-Chief. You may also apply to be an Associate Editor-In-Chief of one of the subtopics below. Please mail us [1] to indicate your interest in serving either as an Editor-In-Chief of the entire topic or as an Associate Editor-In-Chief for a subtopic. Please be sure to attach your CV and or biographical sketch.

Overview
A polyhistidine-tag is an amino acid motif in proteins that consists of at least six histidine (His) residues, often at the N- or C-terminus of the protein. It is also known as hexa histidine-tag, 6xHis-tag, and by the trademarked name His-tag® (registered by EMD Biosciences). The tag was invented by Roche and its vectors are distributed by Qiagen. A Variety of purification kits for histidine-tagged proteins are available from Qiagen, Sigma, Thermo Scientific, GE Healthcare and others.
The use of the tag for academic users is unrestricted; however, commercial users must pay royalties to Roche. Suitable tag sequences are available for free commercial use; for example, MK(HQ)6 may be used for enhanced expression in E. coli and tag removal. The total number of histidine residues may vary in the tag. The his-tag may also be followed by a suitable amino acid sequence that facilitates a removal of the polyhistidine-tag using endopeptidases. This extra sequence is not necessary if exopeptidases are used to remove N-terminal His-tags (e.g., Qiagen TAGZyme). Furthermore, exopeptidase cleavage may solve the unspecific cleavage observed when using endoprotease-based tag removal. Polyhistidine-tags are often used for affinity purification of genetically modified proteins.
Applications
Protein purification
Polyhistidine-tags are often used for affinity purification of polyhistidine-tagged recombinant proteins that are expressed in Escherichia coli [1] or other prokaryotic expression systems. The bacterial cells are harvested by centrifugation and the resulting cell pellet can be lysed by physical means or with detergents or enzymes such as lysozyme. The raw lysate contains at this stage the recombinant protein among several other proteins derived from the bacteria and are incubated with affinity media such as NTA-agarose, HisPur resin or Talon resin. These affinity media contain bound metal ions, either nickel or cobalt to which the polyhistidine-tag binds with micromolar affinity. The resin is then washed with phosphate buffer to remove proteins that do not specifically interact with the cobalt or nickel ion. The washing efficiency can be improved by the addition of 20 mM imidazole and proteins are then usually eluted with 150-300 mM imidazole, but higher concentrations are used as well. The purity and amount of protein can be assessed by SDS-PAGE and western blotting.
Affinity purification using a polyhistidine-tag usually results in relatively pure protein when the recombinant protein was expressed in prokaryotic host organism. In special cases or for special purposes like the purification of protein complexes to study protein interactions, purification from higher organisms such as yeast, insect cell or other eukaryotes may require a tandem affinity purification[2] using two tags to yield higher purity. Alternatively, single-step purification using immobilized cobalt ions rather than nickel ions generally yields a substantital increase in purity and requires lower imidazole concentrations for elution of the his-tagged protein.
Because its mechanism is only dependent on the primary structure of proteins, polyhistidine-tagging is the option of choice for purifying recombinant proteins in denaturing conditions. Generally for this sort of a technique, histidine binding is titrated using pH instead of imidazole binding -- at a high pH histidine binds to nickel but at low pH (~4) histidine becomes protonated and is competed off of the metal ion. Compare this to antibody purification and GST purification which require some protein be properly folded.
Polyhistidine-tag columns will retain several well known proteins as impurities. One of them is FKBP-type peptidyl prolyl isomerase, which appears around 25kDa (SlyD). These should be separated in any protein prep by using a secondary chromatographic technique.
Binding assays
Polyhistidine-tagging can be used to detect protein-protein interactions in the same way as a gst pulldown assay. However, this technique is generally considered to be less sensitive, and also restricted by some of the more finicky aspects of this technique. For example, reducing conditions cannot be used, EDTA and many types of detergents cannot be used.
Adding Polyhistidine Tags
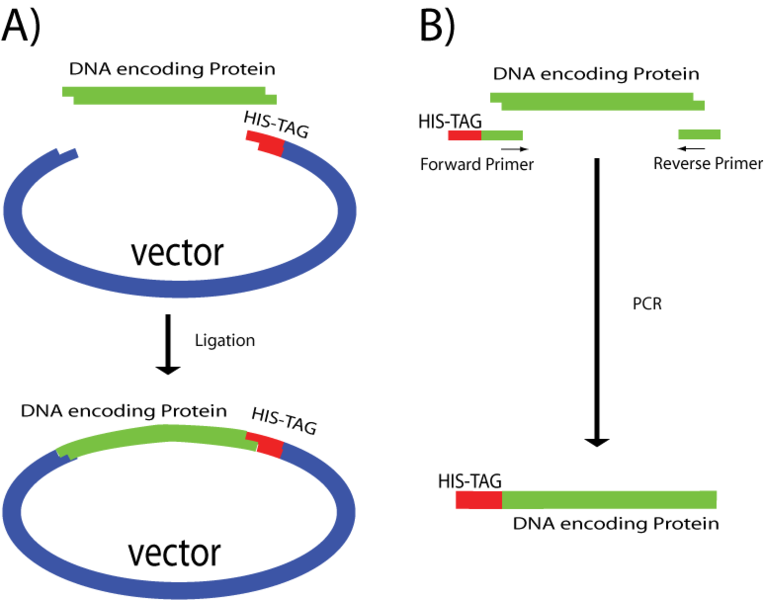
The most common polyhistidine tags are formed of six histidine (6xHis tag) residues which are added at the C-terminal or N-terminal of the protein of interest. The choice of the end where His-tag is added will depend mainly on the characteristics of the protein and the methods chosen to remove the tag. Some ends are buried inside the protein core and others are important for the protein function or structure. In those cases the choice is limited to the other end. On the other hand most available exopeptidases can only remove the His-tag from the N-terminal, therefore removing the tag from the C-terminal will require the use of other techniques.
There are two ways to add polyhistidines. The most simple is to insert a protein DNA in a vector encoding a His-tag so that it will be automatically attached to one of its ends (See picture). Another technique is to perform a PCR with primers that have repetitive histidine codons (CAT or CAC) right next to the START or STOP codon in addition of several (16 or more) bases encoding specifically to the protein to be tagged, see primer example below.
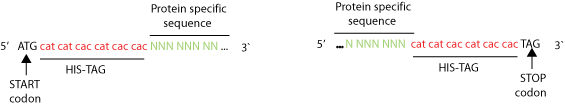
Example of primer designed to add a 6xHis-tag using PCR. Eighteen bases coding six histidines are inserted right after the START codon or right before the STOP codon. At least 16 bases specific of a gene of interest are need it next to the His-tag.
Detection
The polyhistidine-tag can also be used in detection of the protein via anti-polyhistidine-tag antibodies in gel staining (SDS-PAGE) with fluorescently labelled metal ions. This can be useful in subcellular localization, ELISA, western blotting or other immuno-analytical methods.
Immobilization
The polyhistidine-tag can be ideally used for the immobilization of proteins on a surface such as on a nickel or cobalt coated microtiter plate or on a protein array.
References
- ↑ Trends Biochem Sci. 1995 20(7):285-6 Purification of His-Tag fusion proteins from Escherichia coli
- ↑ Nature. 2002 415(6868):141-7 Functional organization of the yeast proteome by systematic analysis of protein complexes
Template:Protein methods Template:Jb1 Template:WH Template:WS